0692
Multi-modal Isointense Infant Brain Image Segmentation with Deep Learning based Methods1Department of Radiology and BRIC, UNC-Chapel Hill, USA, Chapel Hill, NC, United States, 2Department of Computer Science, UNC-Chapel Hill, USA, Chapel Hill, NC, United States
Synopsis
Purpose
Accurate segmentation of infant brain images into different regions of interest is very important in studying early brain development. In the isointense phase (i.e., approximately 6-8 months of age), white matter (WM) and grey matter (GM) exhibit similar levels of intensities in Magnetic Resonance (MR) images, due to the ongoing myelination and maturation. This results in extremely low tissue contrast and thus makes tissue segmentation very challenging. Existing methods for tissue segmentation in this isointense phase usually employ patch-based sparse labeling on single modality. In this paper, we propose a 3D fully convolutional network trained in an end-to-end and voxel-to-voxel fashion for segmentation of isointense-phase brain images. To better model small brain structures, we extend the conventional FCN architectures from 2D to 3D, and also, rather than directly using FCN, we further integrate coarse (highly-semantic) and dense (naturally high-resolution) feature maps to better model small tissue regions. To avoid the signals from deep layers being ignored, we further propose to use an additional convolutional layer to make the number of channels similar. More importantly, we adopt a batch normalization strategy to boost the potential optimization problems, making the model converge at a much faster speed, while still increasing the discrimination capability.Method
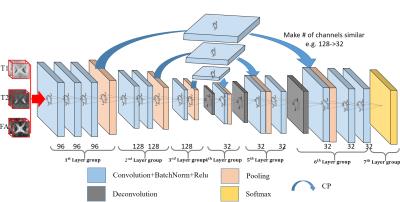
As described by [1], FCN consists of two major operations: down-sampling and up-sampling. The down-sampling operation streams (i.e., convolution and pooling) usually result in coarse and global predictions based on the entire input of the network; and the up-sampling streams (i.e., deconvolution) can generate dense prediction through finer inference. We know that higher-layer features induce higher abstraction of the input, and therefore they can easily ignore small-structure tissues in the 3D image. To solve this issue, we propose to include a pass-through operation in our architecture, in which we take advantage of the context information from the coarse feature maps of the down-sampling set of operations, by copying and fusing the whole learned feature maps to the up-sampling layers. This is similar to the operations used in Long et al. [1], U-Net [2], and Eigen et al. [3]. In this way, we pass the context information from coarse feature maps through the network and use them in the up-sampling phases. As a consequence, the convolution layers during the up-sampling phase can generate more precise outputs based on the assembled feature maps. Furthermore, as the number of feature maps for the two combined layers usually differs a lot (i.e., the original deeper layers usually have much less channels than the shallower layers), the signals from the deeper layers can be ignored during the fusion operation. Thus, we propose to use an additional convolutional layer to adjust the number of feature maps from lower layers to be comparable to the corresponding higher layer (Note, this additional layer will not change the size of the feature maps, just adjust the number of them). This architecture is shown in Fig. 1. Furthermore, a batch normalization [4] operation is adopted after each convolution operation, to make the network easier to converge.Results
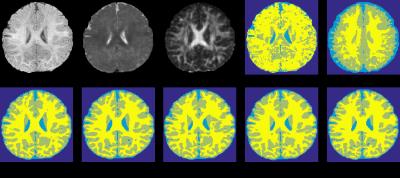
We conduct experiments on a dataset of 11 subjects, each containing 3 imaging modalities (T1, T2 and FA), in a leave-one-out cross-validation fashion. To qualitatively demonstrate the advantage of the proposed method, we first show the segmentation results of different tissues for a typical subject in Fig. 2.
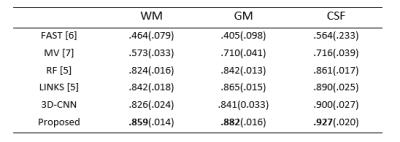
To quantitatively evaluate the segmentation performance, we use Dice ratio to measure the overlap between automated and manual segmentation results. We report the segmentation performance in Table 1. We can observe that our proposed method outperforms other approaches (p=0.0369). Specifically, our method can achieve average Dice ratios of 0.9269 for CSF, 0.8817 for GM, and 0.8586 for WM. In contrast, one of the state-of-the-art methods, i.e., Random Forest with auto-context model (LINKS) [5], achieved the overall Dice ratios of 0.8896, 0.8652, and 0.8424 for CSF, GM and WM, respectively, showing the superiority of our proposed approach.
Conclusion
In this paper, we have proposed a deep learning based method to segment isointense-phase brain images with multi-modality MR images. In our designed deep learning model, we integrate coarse layer information with dense layer information to refine the segmentation performance. Furthermore, we propose to employ batch normalization to make the networks converge faster and better. We have compared our proposed method with several commonly-used segmentation approaches, and the experimental results show that our proposed method outperforms all comparison algorithms on isointense-phase brain segmentation, in terms of both segmentation accuracy. In addition, our work also presents a novel way of fusing multi-layer information to better conduct brain tissue segmentation.Acknowledgements
This work was supported in part by the National Institutes of Health grants MH100217, MH070890, EB006733, EB008374, EB009634, AG041721, AG042599, and MH088520.References
[1] Long, J., E. Shelhamer, and T. Darrell, Fully convolutional networks for semantic segmentation. arXiv preprint arXiv:1411.4038, 2014.
[2] Ronneberger, O., P. Fischer, and T. Brox. U-net: Convolutional networks for biomedical image segmentation. in International Conference on Medical Image Computing and Computer-Assisted Intervention. 2015. Springer. [3] Eigen, D., C. Puhrsch, and R. Fergus. Depth map prediction from a single image using a multi-scale deep network. in Advances in neural information processing systems. 2014. [4] Ioffe, S. and C. Szegedy, Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167, 2015. [5] Wang, L., et al., LINKS: learning-based multi-source IntegratioN frameworK for Segmentation of infant brain images. Neuroimage, 2015. 108: p. 160-72. [6] Woolrich, Mark W., et al. Bayesian analysis of neuroimaging data in FSL. Neuroimage 45.1 (2009): S173-S186. [7] Iglesias, Juan Eugenio, and Mert R. Sabuncu. Multi-atlas segmentation of biomedical images: a survey. Medical image analysis 24.1 (2015): 205-219.Figures