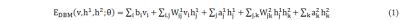0691
Deep Boltzmann Machines-Driven Method for In-treatment Heart Motion Tracking Using Cine MRI1Department of Radiation Oncology, Washington University in St. Louis, St. Louis, MO, United States, 2Laboratoire LITIS, University of Rouen, France, 3Department of Biomedical Engineering, Washington University in St. Louis, St. Louis, MO, United States
Synopsis
We developed a hierarchical deep learning shape model-driven method to automatically track the motion of the heart, a complex and highly deformable organ, on two-dimensional cine MRI images. The deep-learning shape model was trained based on a Deep Boltzmann Machine (DBM)1,2 to characterize both global and local shape properties of the heart for accurate heart segmentation on each cine frame. Preliminary experimental results demonstrate the superior shape tracking performance of our proposed method versus two other methods. The tracking method is designed for heart motion pattern analysis during MRI-guided radiotherapy and the subsequent evaluation of potential heart toxicity from radiotherapy.
Purpose
This study aims to automatically track in-treatment heart motion on cine MRI images where the shape and position of the heart can change significantly frame-by-frame due to both cardiac and respiratory motion.Methods
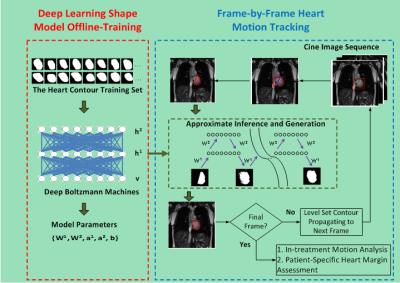
As shown in Fig. 1, the proposed heart tracking method includes two phases:1) deep learning shape model training and 2) frame-by-frame heart motion tracking. In the training phase, we use a three-layered DBM1,2 to train a shape model based on the extracted hierarchical architecture of heart shapes. More precisely, let v denote binary visual units of the visible layer that represents to-be-detect shape, and h1 and h2 represent the binary hidden units of the lower and higher hidden layers. We consider the DBM proposed in literature [1, 2] whose energy function includes three layers and can be represented by Eq. (1) in Fig. 2. Where θ={W1,W2,a1,a2,b} are the model parameters, with W1 and W2 denoting the weights of visible-to-hidden and hidden-to-hidden layers, and a1, a2, and b referring to the biases of h1, h2, and v. In this study, the DBM was trained with a set of 300 heart contours, which were delineated by a radiation oncologist on 360 cine images spanning three patient acquisitions. h1 and h2 of the DBM were trained with 2000 and 100 units respectively. The model parameters {W1,W2,a1,a2,b} were obtained during the training phases and were used for heart motion tracking.
In the tracking stage, a two-step method was designed to extract a heart segmentation on each cine frame. A partial contour was first defined along the left and inferior heart boundaries where large, robust image gradients enable automatic segmentation. In particular, we used a distance-regularized level set method (DRLS)3 to obtain a coarse heart segmentation on each cine frame. The minor diagonal of a bounding rectangle of the segmentation smoothed by a morphological opening operation was used to determine precise left and inferior heart boundaries. The two endpoints of each partial contour were connected to produce a closed contour which was used as the input of the trained DBM-based shape model for approximately inferring the complete heart contour. A layer-wise block-Gibbs sampling scheme2 was utilized to perform approximate inference. After a specific number of iterations were finished, a final binary shape image was produced to provide the heart segmentation result.
Results
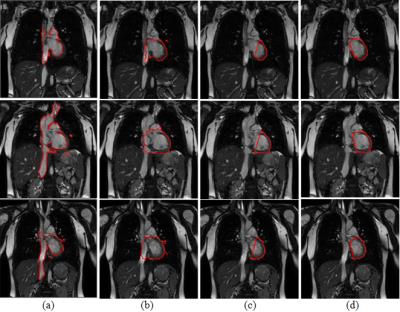
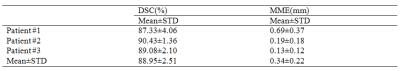
We applied the proposed tracking method on six cine sequences of three patient cases (1 free-breathing and 1 breath-hold for each patient) acquired on a 1.5 T MRI scanner (Philips), with image resolution of 176×176 pixels2, spatial resolution of ~2×2 mm2, and acquisition rate of 3.3 frames/s. The dice similarity coefficient (DSC) and the mean margin error (MME)4 were utilized to evaluate the consistency between the automatic result and the ground truth (i.e. manual contours). As shown in Table 1, the resulting average DSC (89.0%±2.5%) and MME (0.3±0.2 mm) indicated good agreement between the automatic results and the ground truth. In addition, we compared the tracking results with two other deformable model-based segmentation methods (Fig. 3). The experimental results demonstrate the superior performance of our proposed method in application to heart tracking.Discussion
Due to the lack of an effective shape constraint, the Chan-Vese level set model5 yielded undesirable fragmental regions. “Boundary leaking” also occurred because of excessive dependence on regional intensity, as indicated in Fig. 3(a). As shown in Fig. 3(b), the DRLS algorithm ensured stability of the level set evolution and avoided fragmental regions; however, this algorithm was still subject to the “boundary leaking” problem.
In the proposed method, DRLS was used to first capture only the left and inferior boundaries of the heart as a partial contour (Fig. 3(c)). The partial contours were used as the input of the trained deep learning shape model to generate the complete heart contour. By using a three-layered DBM with two hidden layers, the complex structure of the heart could be reliably captured. Even though cardiac and respiratory motion caused significant heart shape changes on different cine images, the proposed method captured heart boundaries, thus yielding accurate segmentation results (Fig. 3(d)).
Conclusion
We presented a motion tracking method that utilizes a deep learning generative shape model and level set propagation scheme to capture heart motion from cine MR images. To the best of our knowledge, this is the first computational method of applying a DBM to model and track the heart shape. The experimental results demonstrate that DBM can accommodate complicated anatomical variation and low image contrast.Acknowledgements
None.References
1. Salakhutdinov R, Hinton GE. Deep Boltzmann Machines. In Proceedings of the 12th International Conference on Artificial Intelligence and Statistics, 2009,448-455.
2. Eslami SM Ali, Heess N, Winn J. The Shape Boltzmann Machine: a Strong Model of Object Shape. International Journal of Computer Vision, 2014, 107(2): 155-176.
3. Li CM, Xu CY, Cui CF, Fox MD. Distance Regularized Level Set Evolution and Its Application to Image Segmentation. IEEE Transactions on Image Processing, 2010, 19(12): 3243-3254.
4. Li H, Chen HC, Dolly S, Li H, et al. An Integrated Model-driven Method for In-treatment Upper Airway Motion Tracking Using Cine MRI in Head and Neck Radiation Therapy. Medical Physics, 2016, 43(8): 4700-4710.
5. Chan TF, Vese LA. Active Contours without Edges. IEEE Transactions on Image Processing, 2001, 10(2): 266-277.
Figures