0637
Time Resolved In Vivo Myofiber Orientations from Combined Cardiac DENSE and cDTI1Department of Bioengineering, University of California, Los Angeles, CA, United States, 2Department of Radiological Sciences, University of California, Los Angeles, CA, United States, 3Biomedical Physics IDP, University of California, Los Angeles, CA, United States
Synopsis
Resolving the dynamics of cardiac myofiber orientations with cDTI could have significant value in the assessment of cardiac disease. Unfortunately, cDTI measurements are not reliable at all cardiac phases and would require extremely long acquisitions. Fusing cDTI with time-resolved displacement maps from cine DENSE could enable evaluation of the dynamics of myofiber orientations using cDTI obtained at a single cardiac phase. The objective of this study was to generate dynamic myocardial fiber maps and validate the approach with dual-phase cDTI measurements.
Background
Cardiac Diffusion Tensor Imaging (cDTI) directly measures myocardial microstructure in vivo. Capturing diffusion tensor dynamics throughout the cardiac cycle has shown value in understanding conditions such as HCM and DCM 1,2. Unfortunately, cDTI is typically limited to measurements at one or two cardiac phases. While recent developments in motion compensated diffusion encoding waveforms have improved cDTI robustness, precise and accurate measurements are not possible at all cardiac phases 3,4. Furthermore, cDTI scans currently take several minutes per cardiac phase, making time-resolved measurements impractical.
Therefore, the objectives of this study were: 1) to generate time-resolved maps of myofiber orientations by combining cDTI data acquired at a single cardiac phase and cine DENSE displacement data; and 2) to validate these time-resolved myofiber maps using information from dual-phase cDTI.
Methods
Imaging: A single mid-ventricular short-axis slice was acquired with cDTI and spiral cine DENSE in healthy volunteers (N=9, 2 females, 7 males, aged 25-42) at 3T (Siemens Prisma) after obtaining informed consent. Imaging parameters for cDTI: free-breathing (navigator triggered) acquired at mid-systole and end-diastole with 2x2x5mm, TE=65ms, TR~3000ms, b-value=0 and 350s/mm2, Navg=10, Ndir=12, scan time=10min; DENSE: balanced 4-point encoding, navigator gated, 2.5x2.5x8mm, TE/TR=1.04/15, Ke=0.06cycles/mm, Navg=3, spiral interleaves=10, scan time=5min with navigator gating.
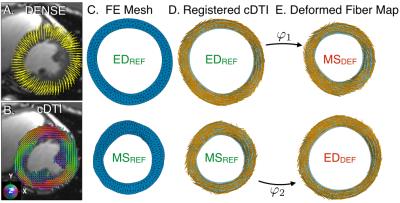
Post-processing (Fig. 1): Lagrangian displacements were extracted from cine DENSE data with custom software 5. cDTI myofiber orientations were co-registered with the DENSE data onto a finite element (FE) mesh of the short-axis slice. Linear displacement interpolation was used to compute the continuum deformation mapping that describes cardiac motion through the cardiac cycle from a chosen reference configuration, here mid-systole (MSREF) or end-diastole (EDREF). The deformation gradient tensor F calculated from φ was used to transform the cDTI myofiber orientations acquired at two times: MSREF and EDREF to their respective computationally deformed configurations at end diastole, EDDEF and mid-systole MSDEF.
This permitted comparing the myofiber maps from the acquired EDREF and MSREF data to the computed EDDEF and MSDEF data to validate the mapping. It was also possible to compare computed fiber maps between methods at any intervening “synthetic” cardiac phase. Mesh elements were directly shared between REF and DEF fiber maps, therefore direct angular comparison between fiber vectors was possible at all phases. Helix angles (HA) were calculated for each mesh element of the EDDEF and MSDEF datasets and were binned by percent wall depth from epicardium (0%) to endocardium (100%) in increments of 10%. Transmural HA distributions were compared between maps at the mid-systolic and end-diastolic time points.
Results
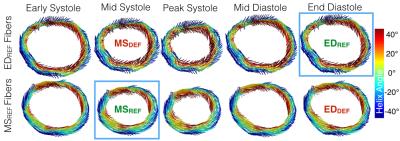
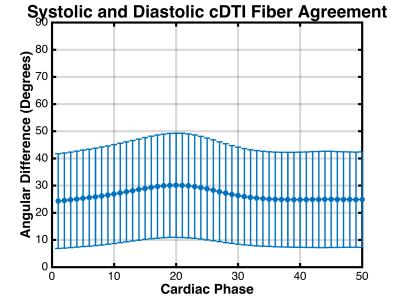
Figure 2 shows the time-resolved myofiber vector maps deformed from both MSREF and EDREF. There was a mean angular difference of 27.2±17.5° between vectors across all phases. Between all myofibers acquired at EDREF and those computed at EDDEF there was a 24.8±18.3° difference. Comparing between all elements in the datasets at each “synthetic” cardiac phase, there was an overall 26.5±18.5° difference between fiber vectors computed with the MSREF and EDREF data (Figure 3).
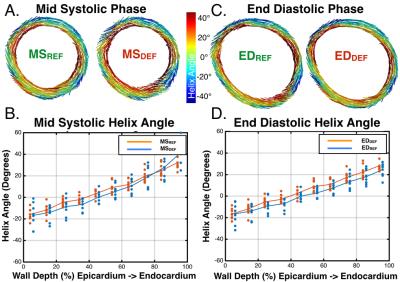
Figure 4 shows transmural HAs across all volunteers, and representative myofiber vector maps colored by HA at the MSREF or EDREF acquisition time points. HA maps were largely in agreement between the two methods. Across all elements in the the mid-systolic (MSREF vs. MSDEF) or end-diastolic (EDREF vs. EDDEF) slices, HA’s differed by (Δθ=9.1±26.3° and 7.7±25.1°), respectively.
Discussion
Combining cDTI and cardiac DENSE data allows for time-resolved maps of myofiber orientation that cannot currently be easily acquired with cDTI alone. Myofiber maps derived independently from systolic and diastolic cDTI data show moderate agreement between fiber angles and better agreement between helix angle maps. Fiber vector agreement was within the cone of uncertainty 6 (data not shown), which measures the precision of cDTI fiber angle measurements, for diastolic phase cDTI measurements.
The difference between time-resolved fiber vectors computed with MSREF and EDREF datasets is greatest during systole. This may arise from the lack of LV twist in the displacement mapping model, which is greatest in systolic time points.
While HA increased transmurally, these values were lower than expected at the epicardial and endocardial border. This could be explained by the interpolation scheme used to register the cDTI fibers to the finite element mesh and conservative masking.
The comparison between MSREF and EDREF mappings validates our model by demonstrating that fibers from one acquired cDTI phase can be deformed through the cardiac cycle using DENSE to another time point within the precision expected for diastolic cDTI itself.
Conclusion
Combining single phase cDTI and cardiac DENSE allows the generation of time-resolved fiber orientations throughout the cardiac cycle with precision near to acquisition precision.Acknowledgements
Funding support from the Department of Radiological Sciences, NIH R01HL131975, and the Center for Duchenne Muscular Dystrophy at UCLA.References
1. Ferreira PF, Kilner PJ, McGill L-A, et al. In vivo cardiovascular magnetic resonance diffusion tensor imaging shows evidence of abnormal myocardial laminar orientations and mobility in hypertrophic cardiomyopathy. Journal of Cardiovascular Magnetic Resonance 2014;16(1):1.
2. von Deuster C, Sammut E, Asner L, et al. Studying Dynamic Myofiber Aggregate Reorientation in Dilated Cardiomyopathy Using In Vivo Magnetic Resonance Diffusion Tensor Imaging. Circulation: Cardiovascular Imaging 2016;9(10):e005018.
3. Stoeck CT, Von Deuster C, Genet M, Atkinson D, Kozerke S. Second-order motion-compensated spin echo diffusion tensor imaging of the human heart. Magnetic resonance in medicine 2015.
4. Aliotta E, Wu HH, Ennis DB. Convex optimized diffusion encoding (CODE) gradient waveforms for minimum echo time and bulk motion–compensated diffusion-weighted MRI. Magnetic resonance in medicine 2016.
5. Spottiswoode BS, Zhong X, Hess A, et al. Tracking myocardial motion from cine DENSE images using spatiotemporal phase unwrapping and temporal fitting. IEEE Transactions on medical imaging 2007;26(1):15-30.
6. Jones
DK. Determining and visualizing uncertainty in estimates of fiber orientation
from diffusion tensor MRI. Magnetic Resonance in Medicine 2003;49(1):7-12.
Figures