0635
DeepVentricle: A Fully Convolutional Neural Network for Automating Functional Measurements in Cardiac MR1Arterys, Inc, San Francisco, CA, United States, 2General Radiology, Stanford University School of Medicine, Stanford, CA, 3Radiology, Stanford University School of Medicine, Stanford, CA
Synopsis
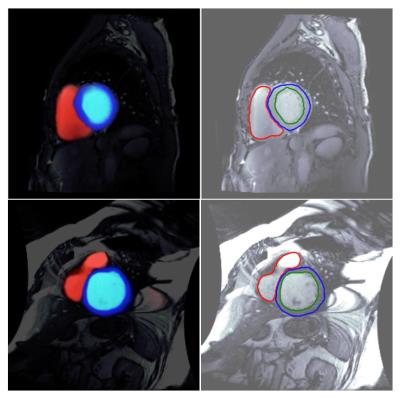
We present DeepVentricle, an automated approach to ventricular segmentation in cardiac MR. DeepVentricle uses a fully convolutional neural network to simultaneously perform semantic segmentation of the left ventricle (LV) and right ventricle (RV) endocardium, and LV epicardium; segmentations are then used to estimate ejection fraction and myocardial mass. We show that the error rates of LV ejection fraction and mass are within the expected range of expert annotator inter-rater variation. This suggests that contours calculated using DeepVentricle could be useful on their own or as an initial estimate for clinicians as part of their semi-automated annotation workflow.
Introduction
Cardiac Magnetic Resonance (CMR) is a technique used to precisely assess cardiac function for patients with known or suspected cardiovascular disease. Although CMR can be used to evaluate cardiac function with great precision, this evaluation traditionally requires a radiologist or technologist to spend tens of minutes per case contouring ventricular structures using manual or semi-automated methods. In this work, we propose DeepVentricle, a fully-automated, deep-learning-based method of contouring the left ventricle (LV) endocardium and epicardium and right ventricle (RV) endocardium, allowing for automated estimates of functional parameters such as ejection fraction and myocardial mass. We show that the error of our method for contouring LV structures is within the expected range of expert inter-rater variation, and that the automated method therefore exhibits performance similar to that of an expert annotator.Methods
Training Data
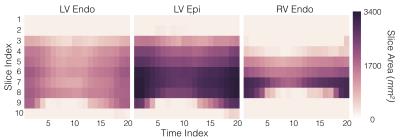
For training the model, we use 898 short-axis cine Steady State Free Precession (SSFP) series from 877 patients. DICOM images and contours were collected as part of standard clinical care at a partner institution. Annotated contour types include LV endocardium, LV epicardium and RV endocardium. A given study may have one or more annotated contour types. Contours were annotated with different frequencies; 100% (897) of studies have LV endocardium contours, 24% (212) have LV epicardium contours and 81% (726) have RV endocardium contours.
Note that, although the RV contours are used in the training process, our current evaluation process benchmarks only the predicted LV contours.
Training Process
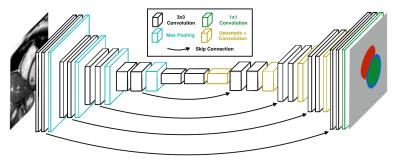
DeepVentricle is a fully-convolutional neural network that is used to semantically segment individual slices from a given CMR study. The network is a variant of the U-Net1 segmentation architecture. Training and inference are performed using the Keras2 deep learning package with TensorFlow as the backend
For each image, the network labels each pixel as one of: (1) background, (2) LV blood pool, (3) LV myocardium or (4) RV blood pool. Because not all ground truth contours are present in every series, we modify the pixelwise cross-entropy loss function to account for missing contours. We discard the component of the loss that is calculated on images for which ground truth is missing; we only backpropagate the component of the loss for which ground truth is known. This allows us to train on our full training dataset, including series with missing contours, without any special treatment of missing contours beyond the modified loss function.
Evaluation
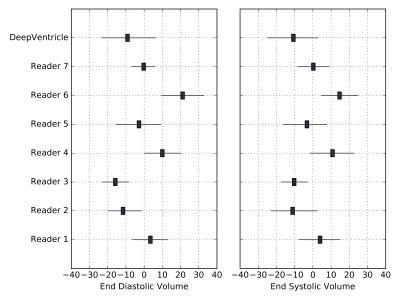
We evaluate our model on on the SCMR Consensus Contour Data Set by Suinesiaputra et al.3, which consists of 15 short-axis CMR studies with contours provided by seven expert annotators. Suinesiaputra et al. defined for each slice of each study a consensus contour using the STAPLE method4, and determined consensus and inter-rater distributions of the following measurements: LV end diastolic (ED) volume, end systolic (ES) volume, ejection fraction (EF) and LV mass (LVM). We compare our error on these four metrics to the inter-rater variation observed among the expert annotators.
Results
We compare (a) our error with respect to the SCMR consensus values from Suinesiaputra et al., averaged over all studies to (b) the standard deviation of expert annotator error, averaged over all studies.
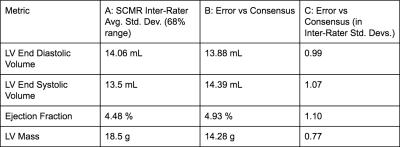
Table 1 summarizes our results. Our average error is close to the average standard deviation of expert annotator error for all metrics.
Discussion
For all measured functional metrics, our automated contouring method achieved an average error that is close to the average standard deviation of expert annotator error. This result suggests that our automated contouring method could potentially be used as a standalone, fully-automated tool for determining CMR functional parameters. Alternatively, our algorithm could provide a reasonable initial estimate of LV endocardium and LV epicardium contours to clinicians for the purpose of calculating ejection fraction and myocardial mass.Conclusion
We present DeepVentricle, an automated approach to ventricular segmentation in cardiac MR. DeepVentricle uses a fully convolutional neural network to simultaneously perform semantic segmentation of the left ventricle (LV) and right ventricle (RV) endocardium, and LV epicardium; LV segmentations are then used to estimate LV ejection fraction and myocardial mass. We show that the error rates of LV ejection fraction and mass are within the expected range of human inter-rater variation. This suggests that contours calculated using DeepVentricle could be useful on their own or as an initial estimate for clinicians as part of their semi-automated annotation workflow.Acknowledgements
We acknowledge NVIDIA for their donation of GPU hardware to support this research effort.References
1. Ronneberger, O., Fischer, P., & Brox, T. (2015, May 18). U-Net: Convolutional Networks for Biomedical Image Segmentation. http://arxiv.org/abs/1505.04597v1.
2. Chollet, F., Keras, (2016), GitHub repository, https://github.com/fchollet/keras
3. Suinesiaputra, A., Bluemke, D. A., Cowan, B. R., Friedrich, M. G., Kramer, C. M., Kwong, R., et al. (2015). Quantification of LV function and mass by cardiovascular magnetic resonance: multi-center variability and consensus contours. Journal of Cardiovascular Magnetic Resonance, 17(1), 63. http://doi.org/10.1186/s12968-015-0170-9
4. Warfield, S. K., Zou, K. H., & Wells, W. M. (2004). Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Transactions on Medical Imaging, 23(7), 903–921. http://doi.org/10.1109/TMI.2004.828354
Figures