0624
Quantitatively Differentiating Multiple Co-Existing Pathologies in High-Grade Glioma1Chemistry, Washington University, St. Louis, MO, United States, 2Neurological Surgery, Washington University, St. Louis, MO, United States, 3Radiology, Washington University, St. Louis, MO, United States, 4Biomedical Engineering, Washington University, St. Louis, MO, United States, 5Pathology and Immunology, Washington University, St. Louis, MO, United States, 6Neurology, Washington University, St. Louis, MO, United States, 7Developmental Biology, Washington University, St. Louis, MO, United States, 8Hope Center for Neurological Disorder, Washington University, St. Louis, MO, United States
Synopsis
We demonstrate diffusion MRI histology (D-Histo) is able to detect, differentiate and quantify various co-existing pathologies and structures including tumor, tumor infiltration, necrosis within human brain tumor specimen while conventional MRI and DTI fails. Quantitative maps of H & E and GFAP were generated and co-registered with D-Histo for voxel-wise correlative analysis. D-Histo-derived restricted-isotropic-diffusion fraction correlated with the area of hematoxylin and GFAP positive stain. D-Histo-derived hindered-isotropic-diffusion fraction successfully predicted the distribution of tumor necrosis corresponding with H & E. D-Histo is promising for brain tumor diagnosis, surgical planning, and treatment response monitoring.
Introduction
MRI is widely used to detect brain tumors and monitor treatment response clinically. More recently, diffusion tensor imaging (DTI) has been applied on pre-surgical planning for brain tumor.1,2 However, tumor-associated structural changes such as vasogenic edema, hemorrhage, necrosis and microvascular proliferation have significantly confounded the sensitivity and specificity of brain tumor detection using conventional MRI or DTI. We have employed a new diffusion MRI histology (D-Histo) approach to distinguish and quantify the extent of tumor cellularity, necrosis and white matter integrity in surgically-resected brain tumor specimens. Different histology staining was quantified to validate D-Histo pathological metrics.Materials and Methods
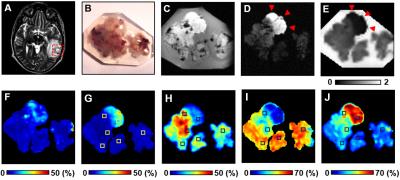
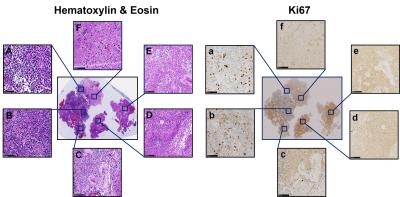
Brain specimen: Six brain tumor specimens (one with anaplastic ependymoma and five with glioblastoma) from five patients were surgically resected and immediately fixed in 10% formalin in phosphate buffered saline at room temperature. Tissues were embedded in agar gel for MR imaging (Fig. 1B) followed by sectioning for Hematoxylin and Eosin (H&E), Ki67 and GFAP staining. Histology slides were digitized using NanoZoomer 2.0-HT System (Hamamatsu, Japan) with a 20× objectives for analyses.
MR imaging and analysis: Specimens were examined using an Agilent DirectDrive console equipped with a 4.7 T magnet and a home-made surface coil using the following parameters: repetition time (TR) 1500 ms, echo time (TE) 40 ms, slice thickness 0.5 mm, field-of-view 2.4 × 2.4 cm2, data matrix 96 × 96. Diffusion sensitizing gradients were applied in 99 directions with max b-value = 3000 s/mm2. Both D-Histo and DTI analyses were performed using an in-house software.
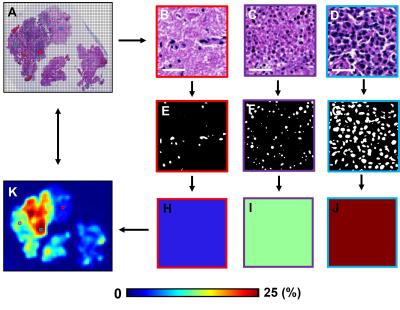
Histology quantification: Raw high-resolution histology image (Fig. 3A) was down-sampled using ImageJ (NIH, Bethesda) NDPI Tools plugin to match MRI voxel size, generating each down-sampled histology image voxel containing 272 × 272 raw histology image pixels (Fig. 3B, C, D). Positive staining was identified and counted in each selected down-sampled histology voxels (Fig. 3E, F, G). The fractions of positive stain area were computed as the ratio between the number of positive staining pixels and the total number of pixels (= 272 × 272) within down-sampled histology voxel (Fig. 3H, I, J). Quantitative histology maps (Fig. 3K) were then generated to correlate with MRI.
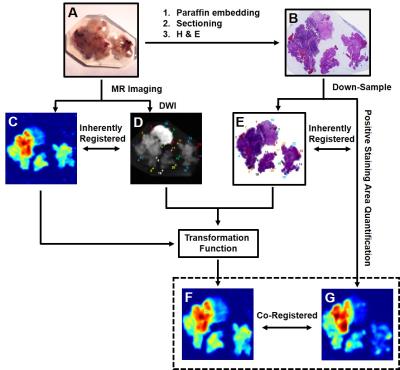
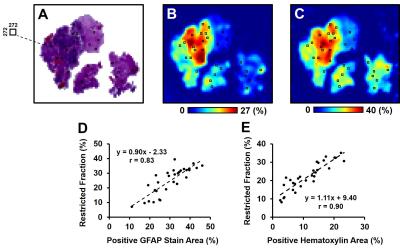
Co-registration and correlation: Transformation function of thin plate spline (TPS) registration was computed in MIPAV (NIH, Bethesda) and applied to warp D-Histo to quantitative histology maps (Fig. 4). Thirty landmark pairs along the perimeter of specimen were manually placed on down-sampled histology images (Fig. 4E) and DW images (Fig. 4F) for TPS transformation. Through successful image co-registration, selected voxels on down-sampled histology images was transferred to MR images. Due to inherently co-registered relations, quantitative positive histology staining was co-registered with D-Histo metrics maps. We randomly selected 30 voxels (Fig. 5A, black squares) from the down-sampled histology image and applied them to the co-registered quantitative histology maps (Fig. 5B) and D-Histo maps (Fig. 5C) for correlative analyses.
Results and Discussions
The widely-accepted notion of tumor signal profile in DWI and DTI, i.e., hyper-intensity in DWI and hypo-intensity in ADC (Fig. 1B, C, red arrows) appeared to be false-positive identification due to complicated pathologies in high grade glioma. Different pathologies or structures were differentiated respectively by individual diffusion profiles through D-Histo analysis. D-Histo-derived restricted-isotropic-diffusion fraction (Fig. 1G) and hindered-isotropic-diffusion fraction (Fig. 1H) maps correctly reflect the distribution of tumor cellularity and necrosis, respectively corresponding to H&E staining (Fig. 2). High restricted-isotropic-diffusion fraction was also associated with high Ki67 staining (Fig. 2H, I). D-Histo-derived anisotropic-diffusion fraction map clearly identified the white matter region (Fig. 1I) where DTI-FA was low and DWI was high (Fig. 1F). D-Histo-derived highly-restricted-isotropic-diffusion fraction map reflected the distribution of glial cells in white matter region (Fig. 2G). Voxel-wise correlations were performed to quantitatively associate D-Histo metrics with individual histopathological components. Restricted-isotropic-diffusion fraction correlated with the area of positive hematoxylin (Fig. 5D; r = 0.90, p < 0.0001) and GFAP staining (Fig. 5E; r = 0.83, p < 0.0001). For other five specimens, restricted-isotropic-diffusion fraction correlated with the area of positive hematoxylin stain (r = 0.78, 0.38, 0.66, 0.51, 0.35; p < 0.0001, < 0.0001, < 0.0001, < 0.0001, < 0.0001, respectively). One specimen (B90) didn’t have a positive GFAP staining thus it was not included for analysis. For the rest of the specimens, restricted-isotropic-diffusion fraction correlated with the area of positive GFAP staining (r = 0.72, 0.55, 0.71, 0.69; p < 0.0001, < 0.0001, < 0.0001, < 0.0001, respectively).Conclusion
D-Histo quantitatively differentiated and quantified multiple coexisting pathologies based on their unique diffusion signatures in high-grade glioma specimens missed by conventional MRI and DTI. D-Histo-derived restricted-isotropic-diffusion fraction correlated with positive hematoxylin and GFAP staining.Acknowledgements
No acknowledgement found.References
1. Golby AJ, Kindlmann G, Norton I, Yarmarkovich A, Pieper S, Kikinis R. Interactive diffusion tensor tractography visualization for neurosurgical planning. Neurosurgery. Feb 2011;68(2):496-505.
2. Wu JS, Zhou LF, Tang WJ, et al. Clinical evaluation and follow-up outcome of diffusion tensor imaging-based functional neuronavigation: a prospective, controlled study in patients with gliomas involving pyramidal tracts. Neurosurgery. Nov 2007;61(5):935-948; discussion 948-939.
Figures