0607
Robust Estimation and In-vivo Validation of the Axon Bundle Diffusivity Profiles1Computer Science, Centro de Investigacion en Matematicas, Guanajuato, Mexico
Synopsis
A stable, accurate and robust-to-noise general framework for the estimation of the intra-voxel axial and radial diffusivity parameters for diffusion-weighted magnetic resonance imaging is presented. The method estimates the diffusion profiles at multi-fiber voxels, improving the estimation of the intra-voxel geometry at challenging microstructure configurations. It naturally constrains the sparsity on the recovered solutions and exploits the spatial redundancy of the axon packs. A useful evaluation metric is proposed: it combines the information of the success rate of the number of bundles and their angular error. A new evaluation method for the in-vivo estimations on large datasets is also proposed.
Introduction and Purpose
The estimation of water molecular displacements from DW-MRI allows inferring the neuronal connections architecture. Spherical Deconvolution (CSD)1 has become very popular because the reasonable number of acquisitions used to recover the fiber orientation distribution function. Some disadvantages are: a) it does not explicitly estimate voxel-wise diffusivity profiles but keeps them fixed. This constant deconvolution response-function assumption is not completely valid even on healthy white-matter (WM)2, and could result in incorrect estimations of the number of diffusion compartments3, b) its inability to disentangle bundle populations on small crossing angles ($$$< 45^{\circ}$$$) and the uncertainty on the estimated orientations4,5. It is important to tackle the drawbacks above in order to the improve estimation of bundle-wise microstructure parameters. Here, we propose a general adaptive framework of the response function at each voxel to estimate diffusion multi-compartment parameters from multi-shell data.Methods
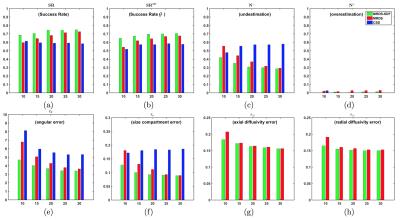
This work illustrates the estimation of the axial and radial diffusivity from a Gaussian Mixture Model (GMM), however our framework can be used for different tissue models. Given a robust estimation of the bundles number and orientations provided by the fixed-response-function MRDS6 we develop a framework to estimate the axial/radial diffusivities $$$\left\{\lambda_{k}^{\|},\lambda_{k}^{\bot}\right\}$$$. MRDS was set to recover a maximum of $$$N=3$$$ tensors per voxel to keep the optimization search space for the diffusivity profiles manageable. This optimization is iterated jointly with the MRDS stage, which re-computes the compartment sizes and orientations from an iteratively-updated response function. To compute the final number of bundles per voxel, the F-Test model-selection is used, which is the recommended selector for the nested models7 on the GMM formulation. The Simultaneous Denoising and Fitting strategy (SDF)6 is also applied; however, to avoid the blurring of local features, $$$\left\{\lambda_{k}^{\|},\lambda_{k}^{\bot}\right\}$$$ are computed only from individual voxel signals. We demonstrate the advantages of combining two widely used performance metrics4,5: Success Rate (SR) and Angular Error ($$$\epsilon_{\theta}$$$), in a more informative metric $$$SR^{\theta}$$$ which quantifies the proportion in which an algorithm estimates the correct number of bundles and their orientations with an angular error $$$< \theta$$$.Gold Standards for the validation on in-vivo data
The proper validation of estimations on in-vivo data is still a challenging open problem. Here we propose a methodology to create robust Gold Standards (GS) from the MASSIVE data set8. We created GSs by using the Bootstrap Aggregating Methodology which reduces the variance of the estimators and the effect of the outliers: it takes a large number of Bootstrap samples, fits a model (CSD, MRDS, etc.) on each sample, and averages the results over all the samples to create a robust estimator. The Bootstrap sampling is a natural option for GS construction on in-vivo data because they present strong outliers due to misalignments, eddy currents, etc. The quality of the GS is verified quantitatively first on synthetic data, then used to evaluate in-vivo results.Experiments and Results
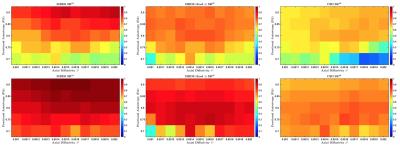
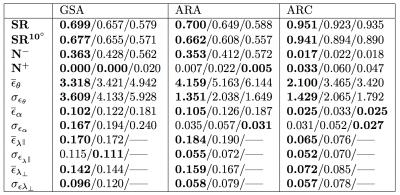
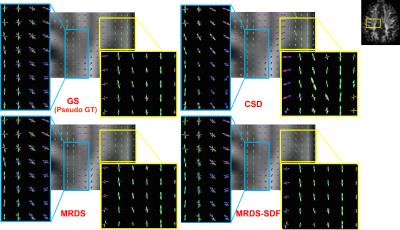
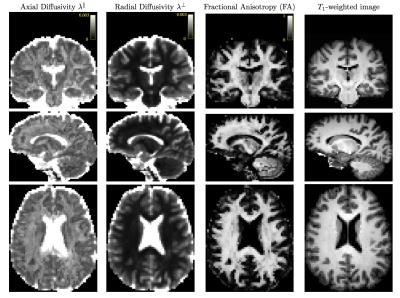
Realistic synthetic MR signals are generated from state-of-the-art multi-compartment models with intra/extra/isotropic compartments9. In all the experiments we use 64 DW signals for $$$b$$$-value = 3000 $$$s/mm^{2}$$$, 16 for $$$b$$$-value = 1000 $$$s/mm^{2}$$$, and 10 $$$S_{0}$$$. We compared against the recent multi-shell CSD10 MRtrix-3 implementation, $$$\ell$$$=8, and $$$th=$$$0.110. The importance of the response function estimation is demonstrated in Figure 1. Results from synthetic experiments on the spatially-coherent phantom in5 are in Figure 2. To build the best as possible GS for in-vivo data, we first create and evaluate GSs for CSD, MRDS and MRDS-SDF for this synthetic phantom using 100 Bootstrap samples. To evaluate the GS-accuracy (GSA) for the three methods, their error with respect to the Ground Truth (GT) of the phantom is computed. We also computed the average realization accuracy (ARA: the errors of the parameters estimated with each method from each sample against the GT), and the average realization consistency (ARC: the errors of the parameters estimated with each method from each sample against their respective GS). The results about the GS’s quality above are presented in Table 1. Based on the evidence from extensive realistic synthetic experiments, we use the MRDS-SDF GS as the best pseudo-GT for in-vivo experiments. The performance of the methodologies w.r.t. this in-vivo pseudo GT is presented in Figure 3. Figure 4 shows slices for the single Bootstrap sample voxel-wise diffusivity profiles computed using MRDS-SDF. MRDS computational time is 9 times larger than CSD’s (voxel-wise).Conclusions
An accurate and robust-to-noise algorithm and a framework for the validation on large in-vivo datasets is presented. It improves the existing approaches in the estimation of the number of axon bundles, their orientation and radial/axial diffusivities at challenging configurations: small crossing angles and different voxel-wise anisotropic diffusion profiles.Acknowledgements
R. Coronado-Leija was supported by a Ph.D. scholarship from CONACYT, Mexico. A. Ramirez-Manzanares and J.L. Marroquin were partially supported by SNI-CONACYT, Mexico, (Grants 169338 and 6243).References
1. Tournier J. D. , Yeh C. H., et al. Resolving crossing fibres using constrained spherical deconvolution: Validation using diffusion-weighted imaging phantom data. NeuroImage. 2008;42(2): 617-625.
2. Gulani V. and Webb A.G., et al. Apparent diffusion tensor measurements in myelin-deficient rat spinal cords. Mag. Reson. in Med. 2001;45(2): 191-195.
3. Parker G.D., Marshall D., et al. A pitfall in the reconstruction of fibre ODFs using spherical deconvolution of diffusion MRI data. NeuroImage. 2013; 65(1): 433-448.
4. Ramirez-Manzanares A., Cook, P. A., et al. Resolving axon fiber crossings at clinical b-values: An evaluation study. Medical Physics. 2011; 38(9): 5239-5253.
5. Daducci A., Canales-Rodríguez E.J., et al. Quantitative Comparison of Reconstruction Methods for Intra-Voxel Fiber Recovery From Diffusion MRI. IEEE Trans. on Med. Imag. 2014;33(2): 384-399.
6. Coronado-Leija R., Ramirez-Manzanares A., et al. Accurate Multi-resolution Discrete Search Method to Estimate the Number and Directions of Axon Packs from DWMRI . Proc. Intl. Soc. Mag. Reson. Med. 23. 2015:565.
7. Burnham K. Model selection and multimodel inference : a practical information-theoretic approach. Springer, New York. 2002.
8. Froeling M., Tax C.M.W., et al. MASSIVE Brain Dataset: Multiple Acquisitions for Standardization of Structural Imaging Validation and Evaluation. Mag. Reson. in Med. 2016; DOI:10.1002/mrm.26259.
9. Panagiotaki E., Schneider T., et al. Compartment models of the diffusion MR signal in brain white matter: A taxonomy and comparison. NeuroImage. 2012; 59(3): 224-2254.
10. Jeurissen, B., Tournier J.D., et al. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. NeuroImage. 2014;103:411-426.
Figures