0418
Investigation of Advanced Diffusion Models in the Spinal Cord: Comparison of NODDI and SMT in MS Patients1Biomedical Engineering, Vanderbilt University, Nashville, TN, United States, 2Vanderbilt University Institute of Imaging Science, Vanderbilt University Medical Center, Nashville, TN, United States, 3Radiology and Radiological Sciences, Vanderbilt University Medical Center, Nashville, TN, United States
Synopsis
NODDI and Spherical Mean Technique (SMT) are multi-compartmental diffusion models that have demonstrated promise in the brain, but have never been applied to the in vivo spinal cord in multiple sclerosis (MS) patients. These models estimate axonal volume fractions, which can be indicative of microstructural integrity. We apply NODDI and SMT in healthy controls and MS patients to evaluate feasibility of both models in the human spinal cord. Results indicate that NODDI (p=0.004) and SMT (p=0.030) can characterize disparity in axonal volume fraction in MS patients, suggesting potential utility of advanced diffusion models in the spinal cord.
Introduction
Spinal cord diffusion tensor imaging (DTI) has
become a powerful tool to assess microstructural changes in multiple sclerosis
(MS). Conventional DTI, however, lacks pathological specificity, and has fueled
a growing interest in the development of more advanced diffusion modeling.
These more advanced models are able to enhance the specificity to individual
diffusion compartments, which can be indicative of microstructural integrity. Specifically,
with Neurite Orientation Dispersion and Density Imaging (NODDI)1,
the intracellular volume fraction is estimated by modeling the orientation
dispersion of neurites, but the complexity of this model requires fixed
intrinsic diffusivities, which is known to vary in white matter tissue. Recently,
the Spherical Mean Technique (SMT)2 has been introduced which
removes the need to estimate fiber orientation distribution, while still
providing an estimate for axonal volume fraction. With SMT, for a fixed b-value, the spherical
mean of the diffusion signal over various gradient directions is independent of
fiber orientation distribution. Therefore, the mean signal over a voxel
represents the microscopic diffusion process.
This study investigates the feasibility of both
NODDI and SMT in the human spinal cord and determines the value of either in
the presence of MS. Methods
Acquisition: Six healthy volunteers (3F/3M, 30.2±5.3 years) participated in this study, four of which had a follow-up scan to assess reproducibility. Five patients with relapsing-remitting MS (5F, 38.6±6.5 years, mean EDSS=2.8) were also recruited. Imaging was performed on a 3T whole body Philips scanner (Philips Achieva, Best, Netherlands) using the two-channel body coil for excitation and a 16-channel SENSE neurovascular coil for reception. The diffusion sequence was acquired in the axial plane and consisted of a cardiac-triggered, reduced field-of-view (FOV), single-shot EPI centered at the C3/C4 level with the following parameters: FOV= 68x52x10 mm3, resolution=1.25x1.25x10 mm3, SENSE (AP)=1.5, TR=3 beats (~3000 ms), TE=65 ms, averages=3. Two shells were acquired: 32 directions at b=711 s/mm2 and 64 directions at b=2855 s/mm2. A series of b=0 s/mm2 images with reversed phase encoding was acquired prior to the scan for post- processing eddy current correction. An anatomical, multi-echo, gradient echo was also acquired (TR/TE1/ΔTE=752/7.1/8.8 ms) for registration and segmentation. The same dataset was used for processing both methods.
Processing: The diffusion-weighted images were corrected for B0 susceptibility and eddy distortion using FSL Topup and Eddy.3 NODDI maps (intracellular volume fraction vin, isotropic volume fraction viso, and orientation dispersion index ODI) were calculated using the freely available UCL NODDI Toolbox; SMT maps (axonal volume fraction vax and axonal diffusivity Dax) were estimated using in-house written code in Matlab.
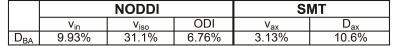
Statistical Analysis: Reproducibility of each metric was assessed using the normalized Bland-Altman difference (DBA), defined as the mean difference of the two scans over the mean diffusion metric, where lower DBA values indicate higher reproducibility. Non-parametric Wilcoxon rank sum test was performed between healthy white matter, lesions and normal appearing white matter (NAWM) to test whether the derived metrics were sensitive to pathological changes.
Results
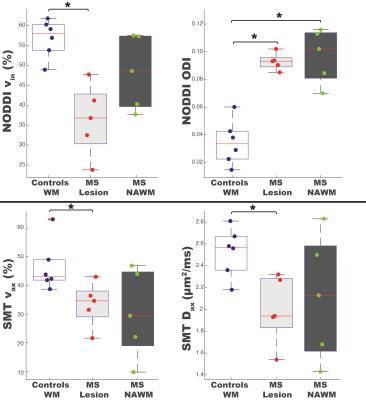
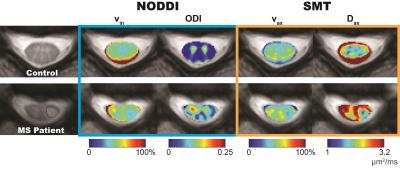
Table 1 indicates that all metrics produce a DBA of 11% or less, demonstrating good agreement between the two scans, except for NODDI’s viso. Notably, while NODDI and SMT both provide metrics for axonal volume fraction, the DBA for SMT’s vax (3.13%) is lower than NODDI’s vin (9.93%), suggesting that SMT’s axonal volume fraction is more reproducible. Figure 1 summarizes the NODDI- and SMT-derived metrics over white matter for healthy controls and MS patients (lesions and NAWM). At the site of the lesion, both NODDI- (p=0.004) and SMT- (p=0.030) derived axonal volume fractions are significantly decreased compared to healthy control white matter. Interestingly, NODDI’s ODI indicates a significant increase in both lesions (p=0.004) and NAWM (p=0.004), suggesting a loss of fiber coherence in pathological tissue. Lastly, SMT shows a marked decrease in Dax (p=0.017) in lesions, as expected with axonal loss. Figure 2 shows the derived maps in an MS patient (EDSS=2, duration of disease=17 years) corroborating the statistical data in Figure 1.Conclusion
Our preliminary results indicate that both NODDI and SMT are capable of distinguishing microstructural changes in the spinal cord in MS. In particular, the methods herein quantify axonal volume fractions in MS, which is not detectable using conventional DTI, suggesting advanced multi-compartmental diffusion models may provide more specific pathological information on characterizing the spinal cord. In comparing NODDI and SMT, SMT shows higher reproducibility in estimating axonal volume fraction and can characterize the decrease of Dax in MS. NODDI, however, provides an estimate of the ODI, which can differentiate white matter in healthy controls from NAWM in MS patients, potentially offering valuable insight in MS prior to lesion formation.Acknowledgements
The authors gratefully acknowledge the following funding sources: DOD W81XWH-13-0073, NIH/NIBIB R21 NS087465-01, NIH/NIBIB R01 EY023240 01A1 and The National MS Society.References
1. Zhang, H., et al., NeuroImage, 2012. 61, p. 1000-16.
2. Kaden, E., et al., NeuroImage, 2016. 139, p. 346-359.
3. Smith, S., et al., NeuroImage, 2004. 23, p. 208-219.
Figures