0400
Fixel-based analysis of Alzheimer's Disease using multi-tissue constrained spherical deconvolution of multi-shell diffusion MRI1Vision Lab, Department of Physics, University of Antwerp, Antwerp, Belgium, 2Computer Imaging and Medical Applications Laboratory, Universidad Nacional de Colombia, Bogota, Colombia, 3Reference Center for Biological Markers of Dementia (BIODEM), University of Antwerp, Antwerp, Belgium, 4Florey Institute of Neuroscience and Mental Health, Melbourne, Australia, 5Department of Radiology, University of Antwerp, Antwerp, Belgium, 6Department of Neurology and Memory Clinic, Hospital Network Antwerp (ZNA) Middelheim and Hoge Beuken, Antwerp, Belgium
Synopsis
In this study, we used multi-shell, multi-tissue constrained spherical deconvolution to investigate group differences in white matter between control subjects, patients with mild cognitive impairment (MCI) due to Alzheimer's Disease (AD) and patients with dementia due to AD. Using the recently proposed fixel-based analysis approach, we distinguish between different fibre populations within a single voxel and characterize them with 3 measures: fibre density, fibre cross-section and the product of these two. We found significant decreases of these metrics in MCI and AD patients compared to healthy controls.
Introduction
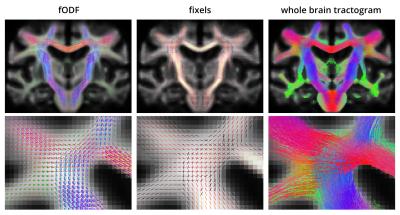
Alzheimer’s disease (AD) is a neurological disorder which is characterized by a decline in cognitive functions and which affects nearly 44 million people worldwide. Previous studies using diffusion MRI (dMRI) have detected significant changes in white matter (WM) pathways in AD patients1. However, most existing studies are difficult to interpret biologically as they are based on voxel-based scalar metrics that are affected to a large extent by partial volume effects (PVE) between adjacent WM bundles. In this study, we employ multi-tissue constrained spherical deconvolution (CSD) of multi-shell diffusion data2 to extract the WM fibre orientation density function (fODF) in each voxel, enabling bundle-specific quantification of WM, even in cases of PVE with adjacent bundles or other tissue types such as gray matter (GM) or cerebrospinal fluid (CSF). Using a recently proposed fixel-based analysis framework, we are able to assess differences in both fibre density (FD) and fibre cross-section (FC) for each fibre element within a voxel (termed ‘fixel’)3.
Methods
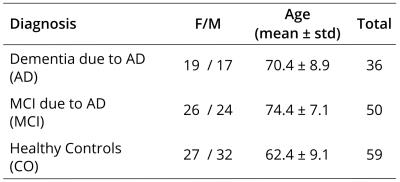
Study population and image acquisition: Patients with MCI due to AD (n=50) and dementia due to AD (n=36), as well as cognitively healthy controls (n=59) were included in the study (see Table 1). All subjects were scanned on a 3T MRI scanner, using a voxel size of $$$2.5\hspace{0.1pt}\times\hspace{0.1pt}2.5\hspace{0.1pt}\times\hspace{0.1pt}2.5\hspace{0.1pt}mm^3$$$. Diffusion weightings of $$$b\hspace{0.1pt}=\hspace{0.1pt}700;\hspace{0.1pt}1000$$$ and $$$2800\hspace{0.1pt}s/mm^2$$$ were applied in 25, 40 and 75 directions, respectively.
Pre-processing: Each data set was pre-processed using a state-of-the-art processing pipeline. Data were first denoised using random matrix theory, thereby increasing the signal-to-noise ratio (SNR) without spatially smoothing the data4. Then, Gibbs-ringing artifacts were suppressed5. Next, combined eddy-current induced distortion and head motion correction was performed using FSL6. Each data set was corrected for bias-fields7 and signal intensities were also normalized across subjects by ensuring that the median signal in pure CSF voxels was constant. Finally, images were up-sampled by a factor of 2 to improve the accuracy of the subsequent spatial normalization.
Multi-tissue CSD: In each subject, we determined a representative dMRI signal response for each of the main tissue types (WM, GM and CSF) using an unsupervised method8. Average tissue responses were obtained across the subjects and then, using the average tissue specific response functions, multi-tissue CSD was performed on each data set to obtain the WM fODF in each voxel (taking into account the presence of GM and CSF).
Spatial normalization: A study-specific fODF population template was constructed from 30 subjects (balanced by group and gender), using a nonlinear diffeomorphic registration algorithm in an iterative atlas building framework9 and all 145 subjects were nonlinearly warped to this template.
Fixel-based measures: From the warped fODFs, we can obtain fixel-wise measures for the fibre density (FD) and cross-section (FC), as well as a combined measure (FDC) obtained by modulating the FD using the FC (FDC=FD×FC)3. FC is able to measure changes in cross-section for a specific fibre bundle based on the volumetric changes when a subject is spatially normalised to the template.
Fibre tracking: Probabilistic whole brain tracking was performed on the template fODFs to determine the long range connectivity between fixels10,11. This information was incorporated into the statistical analysis, a process known as connectivity-based fixel enhancement (CFE)12.
Statistical Analysis: FD, FC and FDC were compared between pairs of groups using permutation tests. Subject age and intra-cranial volume (ICV)13 were included as covariates. For each metric, we highlight the fixels for which $$$p-value\hspace{0.1pt}<\hspace{0.1pt}0.05$$$ after family-wise error correction.
All steps were performed using MRtrix311, unless stated otherwise.
Results
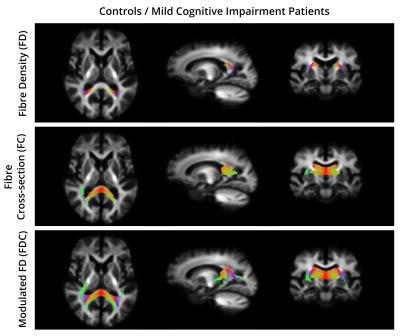
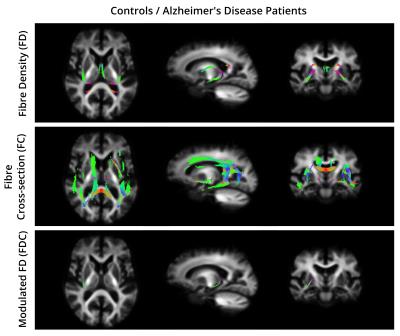
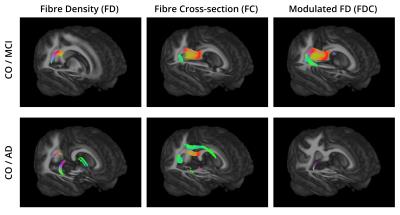
For all the comparisons, the fixels that exhibited a significant decrease in FD, FC, and FDC were matched with the streamlines in the template tractogram. Then, a WM atlas14 was non-linearly registered to the template space and overlapped with the highlighted regions to find their anatomical labels. Figure 2 and 3 show the WM regions with significant decreases in FD, FC and FDC in MCI compared to CO and AD compared to AD, respectively. No significant differences were found when comparing AD to MCI.Discussion and Conclusion
To the best of our knowledge, this is the first study that combines multi-shell multi-tissue CSD and fixel-based analysis to detect WM changes related to AD. Regions of reduced FD, FC and FDC were detected in both AD and MCI subjects. Previous approaches using diffusion tensor or diffusion kurtosis imaging, cannot account for PVEs with adjacent fiber bundles or tissue types. In contrast, fixel-based metrics obtained with multi-tissue CSD allow to distinguish between fibre populations within a voxel, making it a promising tool for measuring axonal degeneration in AD.
Acknowledgements
BJ is a postdoctoral fellow supported by FWO-Vlaanderen.References
1. H. Struyfs, W. Van Hecke, J. Veraart, J. Sijbers, S. Slaets, M. De Belder, L. Wuyts, B. Peters,K. Sleegers, and C. e. a. Robberecht. Diffusion kurtosis imaging: A possible mri biomarkerfor ad diagnosis? Journal of Alzheimer’s Disease, 48(4):937–948, 2015.
2. B. Jeurissen, J.-D. Tournier, T. Dhollander, A. Connelly, and J. Sijbers. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. NeuroImage, 103:411 – 426, 2014.
3. D. A. Raffelt, J.-D. Tournier, R. E. Smith, D. N. Vaughan, G. Jackson, G. R. Ridgway,and A. Connelly. Investigating white matter fibre density and morphology using fixel-basedanalysis. NeuroImage, pages –, 2016.
4. J. Veraart, E. Fieremans, and D. S. Novikov. Diffusion MRI noise mapping using random matrix theory. Magnetic Resonance in Medicine, 76(5):1582-1593, 2016.
5. E. Kellner, B. Dhital, V. G. Kiselev, and M. Reisert. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magnetic Resonance in Medicine, 76(5):1574-1581, 2016.
6. J. L. Andersson and S. N. Sotiropoulos. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage, 125:1063 – 1078, 2016.
7. N. J. Tustison, B. B. Avants, P. A. Cook, Y. Zheng, A. Egan, P. A. Yushkevich, and J. C. Gee. N4itk: Improved N3 bias correction. IEEE Transactions on Medical Imaging, 29(6):1310–1320, 2010.
8. T. Dhollander, D. Raffelt, and A. Connelly. Unsupervised 3-tissue response function estimation from single-shell or multi-shell diffusion mr data without a co-registered T1 image. ISMRM Workshop on Breaking the Barriers of Diffusion MRI, 2016.
9. D. Raffelt, J.-D. Tournier, J. Fripp, S. Crozier, A. Connelly, and O. Salvado. Symmetric diffeomorphic registration of fibre orientation distributions. NeuroImage, 56(3):1171 – 1180, 2011.
10. R. E. Smith, J.-D. Tournier, F. Calamante, and A. Connelly. Sift: Spherical-deconvolution informed filtering of tractograms. NeuroImage, 67:298 – 312, 2013.
11. J.-D. Tournier, F. Calamante, and A. Connelly. Mrtrix: Diffusion tractography in crossing fiber regions. International Journal of Imaging Systems and Technology, 22(1):53–66, 2012.
12. D. A. Raffelt, R. E. Smith, G. R. Ridgway, J.-D. Tournier, D. N. Vaughan, S. Rose, R. Hender-son, and A. Connelly. Connectivity-based fixel enhancement: Whole-brain statistical analysis of diffusion MRI measures in the presence of crossing fibres. NeuroImage, 117:40 – 55, 2015.
13. I. B. Malone, K. K. Leung, S. Clegg, J. Barnes, J. L. Whitwell, J. Ashburner, N. C. Fox,and G. R. Ridgway. Accurate automatic estimation of total intracranial volume: A nuisance variable with less nuisance. NeuroImage, 104:366 – 372, 2015.
14. S. Mori, K. Oishi, H. Jiang, L. Jiang, X. Li, K. Akhter, K. Hua, A. V. Faria, A. Mahmood,R. Woods, A. W. Toga, G. B. Pike, P. R. Neto, A. Evans, J. Zhang, H. Huang, M. I. Miller,P. van Zijl, and J. Mazziotta. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage, 40(2):570 – 582, 2008.
Figures