0374
Difference Image Ultra-Short Echo Time T2* Mapping Using a 3D Cones Trajectory1Electrical Engineering, Brigham Young University, Provo, UT, United States, 2University of California San Diego, San Diego, CA, United States, 3Brigham Young University, Provo, UT, United States, 4Electrical Engineering, Brigham Young University, 5Radiology, University of Utah
Synopsis
This study introduces a methodology for detecting subtle variations in tissues with very rapid T2* decay through a difference image ultra-short T2* mapping technique using a 3D cones k-space trajectory. The new method is demonstrated in both a normal and surgically repaired Achilles tendon. The resulting UTE images were differenced and T2* values were calculated using a mono-exponential least squares fit on a voxel by voxel basis. The ultrashort T2* maps yield very consistent short T2* values in healthy tendon of 0.3 – 0.5 ms, while notable variations and elevations of T2* values are observed in the surgically repaired tendon.
Introduction
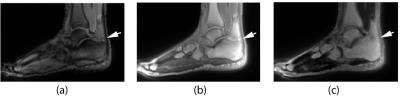
A variety of tissues in MRI are invisible using traditional pulse sequences due to their extremely rapid transverse signal decay. This rapid decay may be caused by a variety of factors, including a short intrinsic T2 relaxation time, but also sub-voxel variations in the magnetic field and other non-idealities. In situations where such extremely rapid signal decay occurs, a UTE sequence must be used to image those tissues. If an image with relatively long TE is subtracted from an image with UTE, then only tissues with extremely short relaxation times remain. An example of a difference image in the ankle is shown in Figure 1. If a series of these images is acquired using a range of TE values, a map of the T2* relaxation times in the tissue of interest may be created.Materials and Methods
Image Acquisition
All images were acquired using a 3T Siemens TIM Trio system using an 8 channel combination foot knee coil. A 3D cones pulse sequence is used because it makes efficient use of gradient capabilities, maximizing gradient amplitude and slew rate, and is very SNR efficient1. A hard RF excitation pulse with 160 ms duration was used with a flip angle of 10°. TR was 10 ms with TE values of 0.25, 0.5, 0.75, 1, and 5 ms for the series of images. FOV was 220x220x220 mm with 1 mm isotropic voxels. Readout time was 2 ms, the minimum possible to reduce T2 blurring while still providing adequate SNR for relaxometry measurements. Image reconstructions were conducted offline using MATLAB.
UTE T2* Mapping
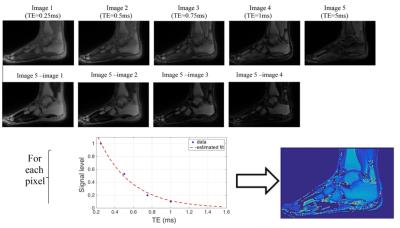
After acquiring the images, post processing was performed on magnitude images in the following way. 1)A set of difference images was formed by subtracting the longest TE image from the shorter TE images. 2)The first difference image was thresholded and used to create a binary mask around the tissue of interest. 3)All of the difference images were multiplied by the binary mask to reduce computation time for determining relaxation times. 4)For each remaining non-zero voxel in the volume of interest, the signal intensities from each difference images were paired their corresponding TE value and a mono-exponential least squares fit was performed. This process, illustrated in Figure 2, was used to compute UTE T2* maps of the ankle in both a healthy volunteer and a volunteer who had an ankle synovectomy.
Results and Discussion
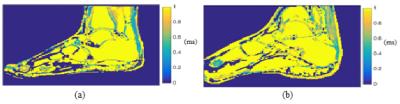
Figure 3 shows the T2* maps comparing a healthy (a) and surgically repaired (b) Achilles tendon windowed to the same scale for comparison. The T2* values in the healthy volunteer across the tendon demonstrate excellent uniformity, providing some confidence in the estimates. Values in the healthy tendon range from 0.32 ms to 0.52 ms. Values in the injured tendon range from 0.5 to 0.74 ms, a notable difference that is easily detected when compared to the healthy tendon.Conclusion
Preliminary data from healthy and unhealthy Achilles tendon demonstrate this methodology can produce excellent in-vivo images of tissues, such as Achilles tendon, with extremely rapid T2* decay. Further, by acquiring UTE T2*maps of these tissues and differencing the images in the manner outlined, an accurate mapping of the T2* values within an unhealthy tendon can be determined. This tool may be extremely useful in characterizing certain types of injuries to tendon and other similar tissues with relatively short T2* values.Acknowledgements
We would like to acknowledge NIH R01EB002524 for providing financial support.References
1Gurney PT, Hargreaves BA, Nishimura DG. Design and analysis of a practical 3D cones trajectory. Magn Reson Med 2006;55(3):575-582.Figures