0331
Identification of potential biomarkers for Parkinson’s disease by 1H NMR spectroscopy1Department of NMR and MRI Facility, All India Institute of Medical Sciences, New Delhi, India, 2Department of Neurology, All India Institute of Medical Sciences, New Delhi, India, 3Department of Biostatistics, All India Institute of Medical Sciences, New Delhi, India
Synopsis
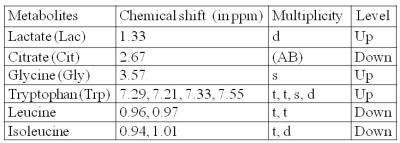
We studied the metabolic profile of urine samples of patients with Parkinson’s disease (PD) and healthy controls (HC) using 700 MHz NMR spectrometer (Varian, M/s Agilent Technologies, USA). The data were processed using Vnmrj (version:2.3A) and binning data estimated using MestReNova software (version :10.0,Mestrelab Research, Spain). PLS-DA multivariate analysis was carried out using MetaboAnalyst (ver.3.0), a web-based metabolomics data processing tool to evaluate significance of metabolites in PD with respect to HC. We observed elevated levels of lactate, tryptophan, glycine and reduced levels of citrate, leucine, isoleucine (t-test, p<0.05), suggestive of several metabolic abnormalities, as mitochondrial dysfunction and reduced bioenergetics efficiency in PD patients.
Purpose
To identify possible biomarkers and understand the pathogenesis of Parkinson’s Disease using NMR techniques.Introduction
The pathological assay of Parkinson’s disease indicate distinct loss of dopaminergic neurons in the substantia nigra pars compacta (SNc), causing the depletion of dopamine in the striatum1. NMR-based metabolomics is a non-invasive technique to study metabolic profiles in bio-fluids and the metabolites associated with physiological and pathological state of the patients. Urine metabolomics is a useful tool for understanding metabolic pathways and networks in Parkinson's disease2.Materials and Methods
Urine samples (after 12 hours fasting) were collected from 28 subjects with idiopathic PD without dementia (15M/13F, mean age: 56.1 ± 6.78 years) and 10 healthy age-matched control (6M/4F, mean age: 51.5 ± 5.57 years) and stored at -80o C until NMR experiments. For the NMR experiments, we used 400µl urine, 30µl TSP (0.5 mM, as a reference) and 170µl phosphate buffer containing 1 mM sodium azide (to prevent bacterial growth). 1D spectrum with water suppression was acquired using a single 90° pulse with 128 scans and 14 s relaxation delay. The data were processed using the Vnmrj (version:2.3A, M/s Agilent Technologies, USA) and binning data estimated using MestReNova software (version10.0, Mestrelab Research, Spain). PLS-DA multivariate analysis was carried out using MetaboAnalyst (ver:3.0), a web-based metabolomics data processing tool for comparison between PD and HC.Results
A total 22 metabolites were assigned unambiguously in the obtained NMR spectra (shown in representative 1H NMR spectra Figure 1). In comparison to HC, levels of 6 metabolites (Table 1) were significantly different on non-parametric t- test (Wilcoxon rank-sum test, p<0.05) in PD patients. The levels of lactate, tryptophan and glycine were observed to be significantly higher while levels of citrate, leucine and isoleucine were lower in PD patients. PLS-DA plot depicts clear separation between PD and HC (Figure 2).
Discussion
Our study revealed 6 metabolites significantly altered in PD patients with respect to HC. Decreased leucine, isoleucine and citrate may be attributed to abnormalities in protein synthesis and energy production in PD patients3. Since glycine is a product of catabolism of fatty acids, which are associated with mitochondrial fatty acid beta-oxidation, altered glycine level in PD patients may be ascribed to mitochondrial disturbances3. Tryptophan was significantly elevated in patients with PD, suggesting changes in tryptophan catabolites that are related to mitochondrial disturbances and impairment of brain energy metabolism involved in the development of neurodegenerative disease4. Elevated level of lactate may be indicative of mitochondrial dysfunction and reduced bioenergetics efficiency in patients with PD with respect to HC5. Since urine has metabolic contributions from different organs and metabolic processes of body, the probability of finding biomarkers are better, though tracking of the exact metabolism becomes difficult.Conclusion
The study indicates mitochondrial dysfunction and reduced bioenergetics efficiency in PD patients that can be detected by NMR spectroscopy.Acknowledgements
No acknowledgement found.References
1. Hatano, T. et al. Identification of novel biomarkers for Parkinson's disease by metabolomic technologies. Journal of Neurol Neurosurg Psychiatry 2016; 87: 295-301.
2. Quinones MP, Kaddurah-DR. Metabolomics tools for identifying biomarkers for neuropsychiatric diseases. Neurobiol Dis. 2009;35:165–76.
3. Luan, H et al. Scientific Reports 2015; 5:13888.
4. Moroni, F. European Journal of Pharmacology.1999; 375: 87-100.
5. Ozelius, LJ et al. New England Journal of Medicine. 2006; 354: 424-425.
Figures