0308
Brain Tumors Disrupt the Resting-State Connectome1Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, United States, 2Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3Department of Biophysics, The Medical College of Wisconsin, Milwaukee, WI, United States, 4Department of Biomedical Engineering, New Jersey Institute of Technology, Newark, NJ, United States, 5Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, MD, United States
Synopsis
Resting-state functional MRI (rsfMRI) has become indispensable for mapping the changes in ‘connectivity’ between brain regions in a range of diseases including brain tumors. However, the complex interplay between abnormal brain tumor vasculature, tumor blood flow, and cancer cell-induced neurovascular uncoupling can confound the interpretation of resting-state connectivity in patients. Therefore, in this preclinical study we quantified brain tumor-induced changes on resting-state connectivity relative to that in healthy brains, followed by histological validation. RsfMRI revealed that brain tumors alter the resting-state connectome, and histology confirmed that this was largely due to cancer cell-induced disruption of the neurovascular unit.
Purpose
Resting state functional MRI (rsfMRI) has become indispensable for mapping the changes in ‘connectivity’ of spatially distinct brain regions1 in a wide array of disease models including brain tumors2. However, the complex interplay between the abnormal brain tumor vasculature, anomalous tumor blood flow3, and cancer cell-induced neurovascular uncoupling4, can confound the acquisition and interpretation of resting-state connectivity in patients2. Since little is known about how the blood-oxygenation-level dependent (BOLD) rsfMRI signal is modulated by the presence of a brain tumor, in this preclinical study we quantified brain tumor-induced changes on the rsfMRI signal and resting state connectivity, relative to that in healthy brains. Histological validation revealed that brain tumor-induced disruption of the neurovascular unit, as well as brain tumor progression were responsible for the observed in vivo changes in resting state connectivity.Methods
9L-GFP brain tumor cells were orthotopically inoculated into the cortices of SCID mice (n=10) as described in5. Brains of healthy SCID mice (n=10) served as the control group. Animals were imaged in vivo on a 400 MHz Bruker spectrometer using the following sequences after localized shimming: (i) T2w rapid acquisition with relaxation enhancement (RARE), RARE-factor=8, TE = 15.0ms, TR = 3.5s, NA=8, resolution = 0.1mm×0.1mm, 16-24 coronal slices, slice = 0.3mm; (ii) gradient-echo EPI, TE = 8.4ms, TR = 400ms, 110 repetitions, resolution = 0.2mm×0.2mm, 16-24 coronal slices, slice= 0.3mm. Animals were imaged under isoflurane anesthesia with body temperature maintained at 37°C and respiration rate at 40-60bpm. After in vivo MRI, animals were perfused with an intravascular tracer (dextran-TRITC, 70kDa), euthanized, brains excised, fixed, frozen and cut for immunofluorescent labeling of neurovascular unit components, such as the astrocyte marker–glial fibrillary acidic protein (GFAP). A 3D mouse atlas6 was used as a reference for segmenting anatomical regions-of-interest (ROI) using Amira® (FEI Software, OR). ROI include left/right: hippocampus(Hi), neocortex(Neo), olfactory bulb(OB), thalamus(Th), striatum(Str), hypothalamus(Hy), and brainstem(Stem). AFNI software was utilized for all image processing7. EPI data were co-registered to the anatomical data, followed by Gaussian 0.5 mm FWHM spatial filtering and 0.01 Hz high-pass filtering. Resting-state connectivity between ROI pairs was evaluated as the cross-correlation (CC) coefficient between their spatially-averaged BOLD signals. Resting-state maps were obtained through cross-correlation analysis of the average resting-state BOLD time course for a given ROI with all the voxels for each comparison ROI. For group-wise comparisons, CC’s for ROI pairs were averaged across each group. Additionally, the resting-state ‘connectome’ of healthy and tumor-bearing mouse brains were represented by force-directed spatial graphs using the Kamada-Kawai algorithm8. Finally, the power spectrum was computed from the average BOLD time-course for each ROI in the tumor and healthy brain.Results
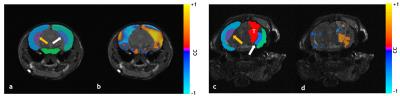
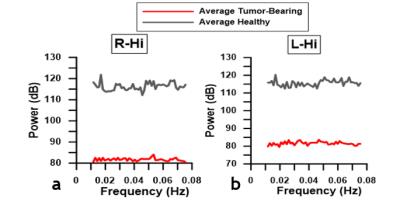
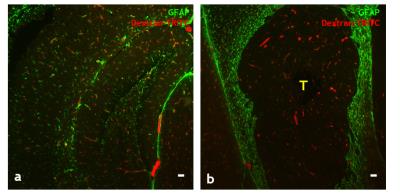
Resting-state maps show that brain tumors significantly alter the resting-state connectivity in the right hippocampus (Fig.1b, 1d). Negative correlations observed in the left hippocampus of the healthy brain (Fig.1b) were attenuated by the presence of a brain tumor (Fig.1d). For healthy brains, ipsilateral ROI exhibited positive correlations, whereas contralateral ROI exhibited negative correlations (Fig.2a). In contrast, connectivity between brain ROI was reorganized for tumor-bearing brains (Fig.2b). Overlaid Kamada-Kawaii plots (Fig.2e) highlight the altered connectome of tumor-bearing brains (Fig.2d) relative to healthy brains (Fig.2c). The global impact of the brain tumor on connectivity was also evident via attenuation of the resting-state BOLD signal between ROI occupying right (Fig.3a) and left hemispheres (Fig.3b). Finally, histology revealed sparse astrocytic-vascular coverage within the brain tumor (Fig.4b) relative to that of healthy brain regions (Fig.4a), thereby validating the presence of neurovascular uncoupling within tumor-associated regions.Discussion
Although rsfMRI has been widely used for presurgical mapping and other applications in brain tumor patients, factors such as cancer cell-induced neurovascular uncoupling can confound the utility of this approach. Recent preclinical rsfMRI tumor studies have primarily focused on BOLD fluctuations arising from abnormal tumor hemodynamics9,10. To the best of our knowledge, this is the first study to characterize the global effect of brain tumor progression on the rsfMRI signal. We discovered that brain tumor-induced alterations in resting-state connectivity were attributable to neurovascular uncoupling as validated by histology. Surprisingly, we observed that progressing brain tumors induced a global reorganization of the resting-state network that extended to the hemisphere contralateral to the tumor, accompanied by global diminution of BOLD signal fluctuations. This work highlights the local and global effects of the brain tumor on its microenvironment, and suggests potential approaches for making rsfMRI a useful cancer biomarker. For example, one could envision ‘normalization’ of the brain tumor vasculature with antiangiogenic therapy, followed by restoration of the ‘brain tumor connectome’ to the ‘heathy brain connectome’ as detected by rsfMRI.Acknowledgements
Supported by NCI 1R01CA196701-01 and 1R21CA175784-02.References
1. Biswal, B. et. al (1995). Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magnetic Resonance in Medicine, 34(4), 537-541.
2. Agarwal, S. et. al (2016). Neurovascular uncoupling in resting state fMRI demonstrated in patients with primary brain gliomas. Journal of Magnetic Resonance Imaging ,43, 620–626.
3. Jain, R. K. et. al (2007). Angiogensis in brain tumors. Nature Neuroscience, 8, 610-622.
4. Lee, Jisook et. al (2009). Glioma-induced remodeling of the neurovascular unit. Brain Research, 1288, 125-134.
5. Pathak, A.P. et. al (2001). MR-derived cerebral blood volume maps: Issues regarding histological validation and assessment of tumor angiogenesis. Magnetic Resonance in Medicine, 46(4), 735-747.
6. Aggarwal, M. et. al (2009). Magnetic resonance imaging and micro-computed tomography combined atlas of developing and adult mouse brains for stereotaxic surgery. Neuroscience, 162(4), 1339-1350.
7. Cox, R. W. (1996). AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Computers and Biomedical Research, 29(3), 162-173.
8. Kamada, Tomihisa and Kawai, Satoru. (1989). An algorithm for drawing general un-directed graphs. Information Processing Letters, 31, 7-15.
9. Baudelet, C. et. al (2006). The role of vessel maturation and vessel functionality in spontaneous fluctuations of T2*-weighted GRE within tumors. NMR in Biomedicine, 19(1), 69-76.
10. Gonclaves, Miguel R. et. al (2015). Decomposition of spontanteous fluctuations in tumor oxygenation usng BOLD MRI and independent component analysis. British Journal of Cancer, 113(8), 1168-1177.
Figures