0256
Predictive cytological topography highlights regions of pathologically confirmed non-enhancing hypercellular tumor in glioblastoma patients1Radiology, Medical College of Wisconsin, Milwaukee, WI, United States, 2Pathology, Medical College of Wisconsin, Milwaukee, WI, United States, 3Neurology, Medical College of Wisconsin, Milwaukee, WI, United States, 4Neurosurgery, Medical College of Wisconsin, Milwaukee, WI, United States
Synopsis
This study combines clinical brain cancer imaging and pathological microscopy with machine learning to generate predictive maps off pathological features (i.e. new contrasts) based on segmented histological cellularity. Predictive cytological topography (PiCT) maps of cellularity were utilized to detect additional pathologically confirmed infiltrative glioblastoma cellularity beyond margins of contrast enhancement.
Purpose
Recent advances in the voxel-wise co-registration of histology obtained from ex-vivo whole brain samples and in-vivo imaging have been used to validate brain cancer imaging biomarkers of hypercellularity and coagulative necrosis1,2. This methodology opens up the possibility of training algorithms to predict histological features based on the MR voxel values and the co-registered histological features of interest.Methods
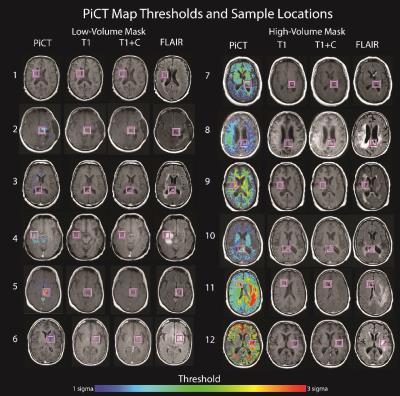
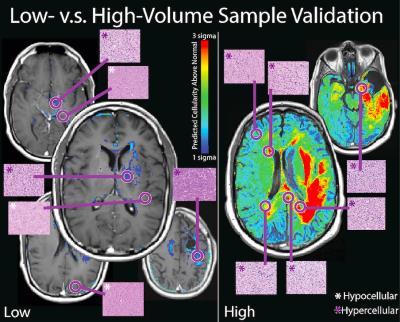
Patient Population Twelve patients with high-grade gliomas were included in this IRB approved study. Patients donated their brains following death. Ex-vivo Histology Processing Tissue samples (approximately 4cm2) were taken from regions suspicious of tumor and free from MR acquisition artifacts in each of the 12 patients. Histological samples were hematoxylin and eosin (H&E) stained. Each slide was then digitized with a motorized microscope stage and Nikon Instruments software photographing tiles at 10x (Melville, NY). Each photo was individually automatically segmented to locate cells using custom Matlab code. Precise Histology to MRI Correlation Co-registration of histology to MRI was performed using a manually defined linear rotational and translation transformation applied to align each histology slide to the MRI. The location of each sample was matched visually to the MRI slice that best represented the sample’s location. Histology from within each region of interest drawn using custom Matlab code was then down-sampled to the MRI resolution for a direct 1 to 1 comparison. Histological segmentation values, along with the MRI values within each voxel were then extracted and combined across all samples. Partial least squares (PLS) regression was applied to the MRI values using cellularity as the independent variable to train a model. The PLS trained model was then applied to the patient’s entire stack of whole brain MR images to generate Predictive Cytological Topographical (PiCT) maps of cellularity (Figure 1). Voxels outside the brain were excluded. The resulting maps were thresholded based on a 95% confidence interval determined from cellularity calculated within a normal histology sample for each patient. To validate the algorithm’s accuracy, additional histology samples were gathered from regions indicated as hypercellular by the PiCT map.Results
Figure 1 shows the PiCT of cellularity for each of the 12 patients. Patients generally fell into two categories, low volume PiCT cellularity and high volume PiCT cellularity. Samples gathered in regions indicated as having a predicted high cellularity all demonstrated viable tumor (Figure 1). Viable tumor was found in regions that appeared normal on conventional imaging in all 12 patients. Figure 2 shows additional regions tested with histology in two patients.Discussion
We present a novel method for mapping brain cancer cellularity with a radio-pathomics approach called predictive cytological topography. This new method may improve surgical planning, radiation guidance, and tumor progression detection.Acknowledgements
Advancing a Healthier Wisconsin, NCI U01-CA176110-01A1References
1. Nguyen HS, Milbach N, Hurrell SL, et al. Progressing Bevacizumab-Induced Diffusion Restriction Is Associated with Coagulative Necrosis Surrounded by Viable Tumor and Decreased Overall Survival in Patients with Recurrent Glioblastoma. AJNR Am J Neuroradiol. 2016.
2. LaViolette PS, Mickevicius NJ, Cochran EJ, et al. Precise ex vivo histological validation of heightened cellularity and diffusion-restricted necrosis in regions of dark apparent diffusion coefficient in 7 cases of high-grade glioma. Neuro Oncol. 2014;16(12):1599-1606.
Figures