0187
PAM50: Multimodal template of the brainstem and spinal cord compatible with the ICBM152 space1NeuroPoly Lab, Institute of Biomedical Engineering, Polytechnique Montreal, Montreal, QC, Canada, 2Montreal Neurological Institute, McGill University, Montreal, QC, Canada, 3Aix-Marseille Univ, CNRS, CRMBM UMR 7339, Marseille, France, 4CEMEREM, Hopital de la Timone, Pôle d’imagerie médicale, AP-HM, Marseille, France, 5Montreal Heart Institute, Montreal, QC, Canada, 6Functional Neuroimaging Unit, CRIUGM, Université de Montréal, Montreal, QC, Canada
Synopsis
Template-based analysis of multi-parametric MRI data of the spinal cord sets the foundation for multi-center studies with minimum bias, thereby helping the discovery of new biomarkers of spinal-related diseases. In this study, we introduce a spinal cord MRI template, the PAM50, which is anatomically compatible with the ICBM152 brain template and uses the same coordinate system. The fusion of the PAM50 and ICBM152 templates facilitates group studies and multi-center studies of combined brain and spinal cord MRI and also allows the use of existing atlases of the brainstem compatible with the ICBM template.
Purpose
Provide a spinal cord MRI template which is anatomically compatible with the ICBM152 brain space.Introduction
Template-based analysis of multi-parametric MRI data of the spinal cord sets the foundation for multi-center studies with minimum bias, thereby helping the discovery of new biomarkers of spinal-related diseases. In a previous study1, a preliminary version of the PAM50 was proposed, which consisted of a T1w, T2w and T2*w MRI template that covered the whole spinal cord and the brainstem and that included atlases of internal structures (white/gray matter, white matter tracts, gray matter sub-regions). However, the referential system of the PAM50 template was not compatible with that from brain templates, preventing the study of the brain-cord axis within a single referential system. The aim of this study was to generate a new version of the PAM50 template which is anatomically compatible and uses the same coordinate system as the ICBM152 space2,3.Methods
Image acquisition: 50 subjects (mean age: 27+-7 y.o., 31/19 men/women) were recruited and scanned in Montreal (n=33) and in Marseille (n=17) on Siemens 3T MRI systems (TIM Trio and Verio, Siemens Healthcare) using the standard head and neck coils. A 3D T1-weighted (T1w) and 3D T2-weighted (T2w) volumes were acquired for each subject. Two FOVs per contrast (1: head and cervical spine; 2: cervical, thoracic and lumbar cord) were acquired and stitched together using off-line software tools provided by the manufacturer’s MRI console after correcting for image bias field. T1w/T2w acquisition parameters were: MPRAGE sequence, TR=2260/1500 ms, TE = 2.09/119 ms, TI = 1200 ms (only T1w), flip angle = 7/140°, bandwidth 651/723 Hz/voxel, voxel size = 1x1x1 mm3. Total acquisition time was 22 minutes.
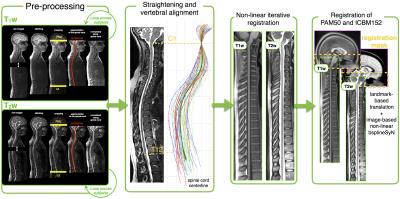
PAM50 generation: The new PAM50 template generation was performed in three steps: (i) the spinal cord centerline and the anterior edge of the brainstem, as well as the intervertebral disks positions were semi-automatically extracted on the T1w and T2w images using the Spinal Cord Toolbox (SCT)4, (ii) the spinal cord (but not the brainstem) was straightened and vertebral levels were aligned using a NURBS-based non-linear transformation and (iii) unbiased left-right symmetric templates were independently constructed using a hierarchical group-wise image-registration method5. Finally, the T1w and T2w templates were co-registered to their respective mid-space and the AMU15 T2*w template6 was merged with the PAM50 template, as described in 5.
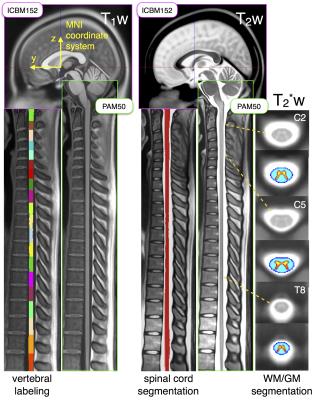
Registration of PAM50 and ICBM152: The fusion of the PAM50 and the ICBM152 templates was performed by registering the brainstem of both templates into a common space (Figure 1). First, a 3D translation transformation was computed between the PAM50 and the ICBM152, using the rostral tip of C1 centered in the cord and the pontomedullary junction as landmarks. Since the average curvature of the brainstem was preserved in the PAM50, brainstem structures in both templates were approximately registered using a 3D translation. Lastly, an image-based iterative non-linear transformation7 was performed to accurately co-register both templates. The physical space of the final PAM50 is the same as the ICBM152 template, thereby providing a common physical coordinates system.
Results
Figure 2 shows the registration of the PAM50 spinal cord template with the ICBM152 brain template. Additionally to being fused with the ICBM152 brain template, the PAM50 covers the spinal cord from C1 to T12, includes the brainstem and part of the brain, is available in T1w, T2w and T2*w MRI contrasts and is registered with atlases of the spinal cord internal structures (i.e, white and gray matter, white matter pathways).Discussion and conclusion
In this study, we propose a referential system that is common for spinal cord and brain MRI studies, by introducing a new version of the PAM50 spinal cord and brainstem template, that is registered with the ICBM152 brain template. The fusion of the PAM50 and ICBM152 templates will facilitates group studies and multi-center studies of combined brain and spinal cord MRI. Existing atlases of the brainstem compatible with the ICBM template could also be used. Moreover, the PAM50 template is available in T1w, T2w and T2*w MRI contrasts and includes atlases of the spinal cord internal structures such as gray/white matter regions and white matter pathways. Future works include adding the lumbar regions into the PAM50 template and validation in patient population.Acknowledgements
This work was supported by the Canada Research Chair in Quantitative Magnetic Resonance Imaging, the Canadian Institute of Health Research [CIHR FDN-143263], the Fonds de Recherche du Québec - Santé [28826], the Fonds de Recherche du Québec - Nature et Technologies [2015-PR-182754], the Natural Sciences and Engineering Research Council of Canada [435897-2013], the Sensorimotor Rehabilitation Research Team (SMRRT), the Functional Neuroimaging Unit (CRIUGM, Université de Montréal), the MEDITIS scholarship program, the PBEEE scholarship program from the Fonds de Recherche du Québec - Nature et Technologies and the Quebec BioImaging Network.References
1. De Leener B, Taso M, Fonov VS, et al. Fully integrated T1, T2, T2*, white and gray matter atlases of the spinal cord. In: Proceedings of the 24th Annual Meeting of ISMRM, Singapore, Singapore. Singapore: 2016 p. 1129.
2. Fonov VS, Evans AC, McKinstry RC, et al. Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. Neuroimage 2009;47:S102.
3. Fonov V, Evans AC, Botteron K, et al. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage 2011;54(1):313–327.
4. De Leener B, Lévy S, Dupont SM, et al. SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data. Neuroimage 2016. DOI: 10.1016/j.neuroimage.2016.10.009
5. Fonov VS, Le Troter A, Taso M, et al. Framework for integrated MRI average of the spinal cord white and gray matter: The MNI-Poly-AMU template. Neuroimage 2014;102P2:817–827.
6. Taso M, Le Troter A, Sdika M, et al. Construction of an in vivo human spinal cord atlas based on high-resolution MR images at cervical and thoracic levels: preliminary results. Magn. Reson. Mater. Phys. Biol. Med. 2014;27(3):257–267.
7. Tustison NJ, Avants BB. Explicit B-spline regularization in diffeomorphic image registration. Front. Neuroinform. 2013;7:39.
Figures