0178
3D Multi-Band, Multi-Slab, and Multi-Shot High-Resolution Diffusion MRI1Brain Imaging and Analysis Center, Duke University Medical Center, Durham, NC, United States, 2Department of Diagnostic Radiology, University of Hong Kong, 3Biomedical Engineering, University of Arizona, Tuscan, AZ, United States
Synopsis
When diffusion MRI data is acquired with 3D multi-slab and/or multi-shot imaging techniques, scan times are often lengthy and phase variations between the acquired shots and/or slice-encoding planes of 3D slabs introduce severe motion artifacts in slice images. To accelerate the acquisition of high spatial resolution diffusion MRI volumes with high SNR and fidelity, we outline a 3D image reconstruction model that simultaneously accounts for both in-plane and through-plane motion artifacts in 3D multi-band, multi-slab and multi-shot diffusion data. Diffusion data acquired and reconstructed in this fashion can be acquired at sub-millimeter spatial resolution with high SNR in ~1-2min.
Purpose:
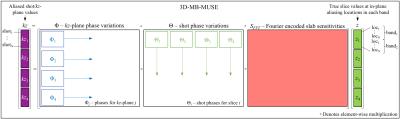
Using single-shot echo planar imaging (EPI) techniques, the spatial resolution of diffusion MRI (dMRI) images is typically constrained by the SNR achievable within the limited acquisition window (due to transverse relaxation time), and the need to minimize motion artifacts and image distortions. With multi-shot EPI, on the other hand, high-resolution dMRI can be reliably achieved through multiplexed sensitivity encoding (MUSE)1, where shot-to-shot phase variations that would otherwise deteriorate image quality are corrected. However, the SNR can be inherently low at very high resolutions. To achieve sufficient SNR, 3D multi-slab imaging techniques can be employed. For single-shot 3D acquisitions, bulk through-plane motion across a 3D slab can be accounted for by using phase variations estimated from low resolution navigator images acquired at kz=0 following the acquisition of each slice encoding plane (kz-plane) of a 3D slab2. In multi-shot 3D acquisitions, however, the time separating the first shot in the first kz-plane in a slab and the last shot in the last kz-plane in the same slab is proportional to TR·Nshots·Nkz, which can span 2-10min for a single dMRI volume and introduce inaccuracy in phase estimation. Accelerating multi-shot 3D acquisitions through multi-band imaging techniques, we present a 3D multi-band MUSE model (3D-MB-MUSE) in Fig. 1, in which Fourier encoded coil sensitivity variations across the excited 3D slabs reconstruct all shots in all kz-planes of a multi-band slab at once, while simultaneously accounting for both in-plane and through-plane motion artifacts across shots/kz-planes/bands.Methods:
Whole brain in-vivo multi-band, multi-slab, and multi-shot dMRI data was acquired in a commercial 3T GE MR750 scanner using a 32 channel head coil. Four interleaved shots were used to image a 25.6cm FOV in a 256×256 acquisition matrix. Custom 3D multi-band pulses simultaneously excited three 10mm slabs spaced 48mm apart. One b=0 and three b=800s/mm2 dMRI volumes were acquired with six multi-band slabs each. Slabs overlapped by 20% and were encoded with 12 kz-planes and 20% oversampling in z, resulting in a spatial resolution of 1.0mm3 with whole-brain coverage. To avoid T1 cross-talk artifacts, odd slabs were acquired in their entirety within a volume before even slabs, thereby enabling the SNR optimal 1.5sec TR for multi-slab imaging3. Under this scheme, the overall acquisition time for each volume was 2min 24sec. In each shot, bulk motion across kz was corrected in the raw data using phase variations estimated from additional low-resolution navigator images acquired at kz=0. Using coil sensitivities derived from an additionally acquired 3D single-band b=0 volume, shot-to-shot and kz-to-kz phase variations were estimated in each of the simultaneously excited slabs using a 3D adaptation of the SENSE4 model before being inserted into 3D-MB-MUSE. Finally, reconstructed slabs were stitched together using slab profile encoding5, and brain extraction was performed using FSL6.Results and Discussion:
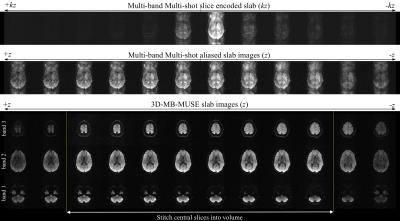
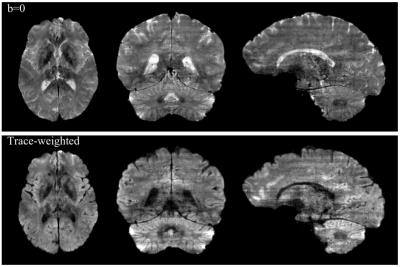
Fig. 2 presents a single diffusion-weighted multi-band multi-shot 3D slab reconstructed with 3D-MB-MUSE. When a 1D-FFT converts the slice encoded multi-shot aliased slab into aliased slab images, severe motion artifacts arise in the phase encoding direction of each slice. By accounting for phase variations across both shots and kz-planes in the three aliased dMRI slabs, 3D-MB-MUSE reconstructs the aliased slice-encoded slab into un-aliased slab images that are free of motion artifacts. While most 3D multi-slab models use coil sensitivities derived from a collapsed slab (acquired at kz=0) to reconstruct all slices in the slab, aliasing artifacts often arise in slices towards the outer edges of thick slabs. Using Fourier encoded sensitivities that vary across the slab, the resulting slab images in Fig. 2 exhibit minimal artifacts from either reconstructing in-plane shots or separating through-plane bands. When stitched together in Fig. 3, the b=0 and trace-weighted volumes exhibit high SNR in 1.0mm3 voxels with only minor slab banding artifacts, which may be further reduced through additional signal-normalization steps in post-processing.Conclusion:
The 3D-MB-MUSE model in Fig. 1 presents an expandable framework that can account for various artifacts associated with 3D multi-slab, multi-band, and multi-shot diffusion imaging. In this abstract, both shot-to-shot and kz-to-kz motion artifacts have been accounted for, and variable coil sensitivities across a slab have been used to separate 3D multi-band slabs acquired with multi-shot EPI. Building on this framework, current work includes accounting for 3D imaging artifacts such as slab profile distortions and T1 cross-talk between slabs, which will ultimately allow slabs to be interleaved within a single 1.5sec TR. Reconstructing interleaved multi-shot and multi-band 3D slabs with 3D-MB-MUSE would permit whole-brain volumes to be acquired at sub-millimeter resolutions with sufficient SNR to produce reliable tractography in ~1min, effectively enabling a widespread adoption of high-resolution dMRI.Acknowledgements
This work was supported in part by NIH grants NS-075017 and R24-106048.References
1. Chen NK, Guidon A, Chang HC, Song AW. A robust multi-shot scan strategy fot high-resolution diffusion weighted MRI enabled by multiplexed sensitivity encoding (MUSE). Neuroimage, 2013; 72:41-47.
2. Engstrom M, Skare S. Diffusion-Weighted 3D multislab echo planar imaging for high signal-to-noise ratio efficiency and isotropic image resolution. Magn. Reson. Med., 2013; 70:1507-1514.
3. Frost R, Jezzard P, Porter DA, Miller KL. Simultaneous multi-slab acquisition in 3D multi-slab diffusion-weighted readout-segmented echo-planar imaging. Proc. Intl. Soc. Magn. Reson. Med. Meeting in Salt Lake City, USA, 2013. Abstract 3176.
4. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity Encoding for Fast MRI. Magn. Reson. Med. 1999; 42:952-962.
5. Van AT, Aksoy M, et al. Slab Profile Encoding (PEN) for minimizing slab boundary artifact in three-dimensional diffusion-weighted multislab acquisition. Magn. Reson. Med. 2015; 73(2):605-613.
6. Jenkinson M, et al. FSL. Neuroimage, 2012; 63:782-790.
Figures