0176
Faster Diffusion-Relaxation Correlation Spectroscopic Imaging (DR-CSI) using Optimized Experiment Design1Electrical Engineering, University of Southern California, Los Angeles, CA, United States
Synopsis
We propose a new experiment design method to accelerate the recent novel diffusion-relaxation correlation spectroscopic imaging (DR-CSI) experiment. DR-CSI acquires imaging data across a range of different b-value and echo time combinations. This enables new insights into tissue microstructure, but the contrast encoding can be slow. Our experiment design approach selects a small subset of the most informative observations to acquire using results from estimation theory. We demonstrate with ex vivo mouse spinal cord MR data that the new experiment design approach enables DR-CSI to be accelerated by a factor of more than 2 without a substantial loss in quality.
Purpose
The diffusion-relaxation correlation spectroscopic imaging (DR-CSI) experiment is a novel high-dimensional extension of traditional diffusion MRI and MRI relaxometry1. Similar to an earlier diffusion-relaxation correlation spectroscopy method (non-imaging)2-3, DR-CSI jointly and nonseparably encodes diffusion and relaxation information, and uses that information to construct a 2D diffusion-relaxation correlation spectrum for every voxel. Compared to traditional analyses, this 2D spectrum enables better resolution of the microstructural tissue compartments that exist inside a macroscopic imaging voxel1. However, despite the potential of DR-CSI, the need to jointly encode both diffusion and relaxation information leads to relatively long acquisition times. In this work, we demonstrate that, guided by principled methods from estimation theory, DR-CSI acquisitions can be substantially accelerated without a substantial loss of information.Theory and Methods
The Cramér-Rao Bound (CRB) is a lower bound on the variance of any unbiased estimator4, and is often used as a metric for deriving optimal experiment designs5-7. We hypothesize that CRB-based experiment design will enable accelerated DR-CSI acquisitions without a significant loss of information. Based on this hypothesis, the CRB for the DR-CSI model is defined by:
$$f(\boldsymbol{\theta},\boldsymbol{\gamma}_n)=\sum_{s=1}^S A_s e^{-b_nD_s} e^{-TE_n/T2_s}$$,
where $$$\boldsymbol{\theta}=\left[ \bf{A}~\bf{D}~\bf{T2} \right]^T \in \mathbb{R}^{3S \times1}$$$ is a set of the unknown diffusion-relaxation parameters to be estimated and $$$\boldsymbol{\gamma}_n = \left[b_n~TE_n \right]$$$ are the experimental conditions for the $$$n$$$th acquisition. Our goal is to choose the experimental conditions $$$\boldsymbol{\gamma}_n = \left[b_n~TE_n \right]$$$ so that the data measurements contain as much information about $$$\boldsymbol{\theta}$$$ as possible. The CRB is defined by $$$\text{COV}(\hat{\boldsymbol{\theta}}) \geq \text{CRB}(\boldsymbol{\theta})=\mathbf{F}^{-1} (\boldsymbol{\theta})$$$, where the Fisher information matrix $$$\mathbf{F}(\boldsymbol{\theta})$$$ is given by $$$[\mathbf{F}(\boldsymbol{\theta})]_{ij} = \sum_{n=1}^N \frac{1}{\sigma^2} \frac{\partial f(\boldsymbol{\theta},\boldsymbol{\gamma}_n)}{\partial \theta_i} \frac{\partial f(\boldsymbol{\theta},\boldsymbol{\gamma}_n)}{\partial \theta_j}$$$ assuming Gaussian random noise statistics. Based on the CRB, the experimental conditions are determined by minimizing the following objective function:
$$J(\boldsymbol{\gamma};\boldsymbol{\theta}) = \sum_{i=1}^{3S}w_i \frac{\sqrt{\left[\text{CRB}(\boldsymbol{\theta})\right]_{ii}}}{\theta_i}$$,
where the $$$w_i$$$ are weighting coefficients to emphasize the interesting tissue parameters.
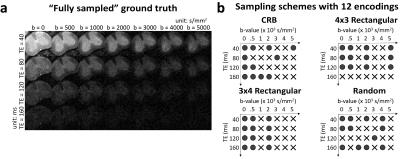
Our experiments used the same ex-vivo mouse spinal cord datasets from previous DR-CSI work1 (three sham controls and three with traumatic spinal cord injury). This data was sampled at every combination of 7 b-values (0, 500, 1000, 2000, 3000, 4000 and 5000s/mm2) and 4 TEs (40, 80, 120 and 160ms) for a total of 28 images. Representative images are shown in Figure 1(a). The spectra estimated from this set of 28 images were considered as a “fully sampled” ground truth.
We used the sequential backward selection algorithm9 to select a subset of 12 contrast-encoding parameters from the original full set of 28. This corresponds to a substantial (2.3x) reduction in experimental duration. For reference, we also compared against 12-encoding rectangular grid and random sampling8 schemes. The sampling schemes are displayed in Fig. 1(b). For rectangular grid sampling, two different options (4 b-values$$$\times$$$3 TEs and 3 b-values$$$\times$$$4 TEs) were considered. For random sampling, ten different sampling realizations were considered, and we report results derived from the best-performing option. For all sampling schemes, DR-CSI spectra were reconstructed using the same dictionary-based spatially-regularized nonnegative least squares optimization approach described in previous DR-CSI work1.
Results and Discussion
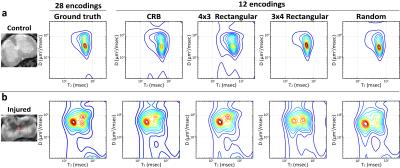
Figure 2 shows spatially-averaged DR-CSI spectra for control and injured spinal cords. In the control cord, the spectra from the CRB-based and 4x3 rectangular grid sampling schemes have the best performance in resolving the two distinct spectral peaks that are known to be present from the “fully sampled” ground truth. In the injured cord, the spectrum from the CRB-based sampling has good consistency with the ground truth, and correctly demonstrates three distinct spectral peaks.
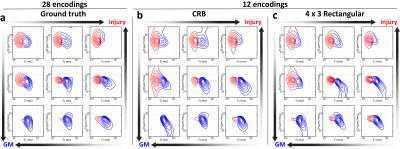
Figure 3(a) shows spatially-varying DR-CSI spectra from a region of the “fully sampled” ground truth in which the tissue transitions between injury and gray matter (GM). Fig. 3(b-c) show the DR-CSI spectra obtained from the CRB-based and 4x3 rectangular grid accelerated acquisitions. The tissue transitions are still seen in the both of the accelerated acquisitions.
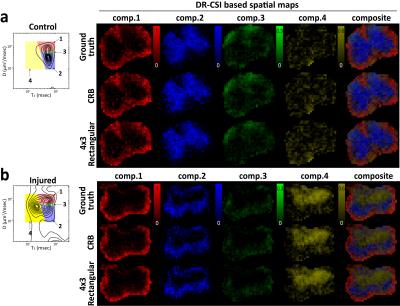
Figure 4 shows that the spatial maps of the integrated spectral peaks from the accelerated scans also have large similarity with the maps from the ground truth. In each case, the maps seem to be consistent with the known anatomy of the spinal cord: The first three components appear to correspond with white matter and gray matter compartments. The last component is only present in the injured cords, and seems to be associated with a new compartment related to the injury.
Conclusions
We demonstrated that DR-CSI can be subsampled, leading to a substantial acceleration of the experiment without a substantial loss of information. Our results also confirm the hypothesis that CRB-based sampling design works well for achieving this acceleration, although other sampling design approaches can also yield similarly good results.Acknowledgements
This work was supported in part by NSF CAREER award CCF-1350563, and NIH research grants R21-EB022951 and R01-NS089212.References
1. Kim D, Kim JH, Haldar JP. Diffusion-relaxation correlation spectroscopic imaging (DR-CSI): An enhanced approach to imaging microstructure. In: Proc. Int. Soc. Magn. Reson. Med. 2016; p. 660.
2. Hürlimann MD, Venkataramanan L, Flaum C. The diffusion-spin relaxation time distribution function as an experimental probe to characterize fluid mixtures in porous media. J Chem Phys 2002; 117:10223-10232.
3. Callaghan PT, Godefroy S, Ryland BN. Use of the second dimension in PGSE NMR studies of porous media. Magn Reson Imaging 2003; 21:243-248.
4. Kay SM. Fundamentals of Statistical Signal Processing, Volume I: Estimation Theory. Upper Saddle River: Prentice Hall, 1993.
5. Cavassila S, Deval S, Huegen C, van Ormondt D, Graveron-Demilly D. Cramér-Rao bounds: an evaluation tool for quantitation. Nuclear Mag Reson Biomed 2001; 14:278-283.
6. Alexander DC, A general framework for experiment design in diffusion MRI and its application in measuring direct tissue-microstructure features. Magn Reson Med 2008; 60:439-448.
7. Zhao B, Haldar JP, Setsompop K, Wald LL. Optimal experiment design for magnetic resonance fingerprinting. In: Proc. IEEE Engineering in Medicine and Biology Conf. 2016; p. 453-456.
8. Benjamini D, Basser PJ. Use of marginal distributions constrained optimization (MADCO) for accelerated 2D MRI relaxometry and diffusometry. J Magn Reson 2016; 271:40-45.
9. Reeves SJ, Zhe Z. Sequential algorithms for observation selection. IEEE Trans. on Signal Processing 1999; 47:123-132.
Figures