4905
Gradient echo modelling with macroscopic field variations and large flip angles1Department of Neurology, Medical University of Graz, Graz, Austria
Synopsis
The signal decay of a 2D gradient echo sequence is substantially influenced by macroscopic field variations along the slice profile. Here we propose a numerical model describing the signal decay due to a macroscopic field gradient for arbitrary excitation pulses with large flip angles. Phantom and in-vivo experiments show that accurate modelling requires inclusion of the phase along the slice profile and the polarity of the slice selection gradient. Additionally, we show that applying the model yields better results for R2*-mapping and myelin water fraction estimation.
Introduction
In the presence of a varying macroscopic field Δω(z), intravoxel dephasing along the slice profile yields an overestimated effective transverse relaxation rate R2*1–6. Based on the Fourier approximation of the slice profile an accurate modeling for gradient echoes with flip angles α<60° was presented in3. Here, we investigated the signal behavior for α>60° for applications that require long TR and thus larger α and propose a numerical model for arbitrary excitation pulses with linear approximated Δω(z).
We show that for a proper modeling the polarity of the slice selection gradient Gslice and the phase along the slice profile have to be considered and demonstrate that not only R2*-mapping but also clinical interesting applications such as whole brain myelin water fraction (MWF) imaging can be substantially improved with the proposed model.
Methods
In a 2D gradient echo sequence the measured signal at time t for slice position zi in slice selection direction z is described by:
$$S(z_{i}, t)=\int_{-\infty}^\infty|M_{xy}(z-z_i)|~e^{i\phi_{xy}(z-z_i)}~e^{i\gamma\Delta \omega(z)t}~ e^{i\phi_0(z)}~dz~S_{model}(t)$$
where |Mxy(z- zi )| is the magnitude and φxy(z- zi) the phase of transverse magnetization, φ0(z) the phase offset and Smodel(t) the signal model. Rewriting Equation (1) by introducing F(zi,t), which describes all components causing additional dephasing to Smodel(t), leads to5:
$$S(z_{i}, t)=~S_{model}(t)~F(z_i,t)$$
Numerical model F(zi,t): |Mxy(z)| and φxy(z- zi) for the RF-pulse envelope B1(t) were estimated with a numerical Bloch solver7. To account for B1+ field variations, B1(t) was multiplied with values obtained from B1-map.To model variations of the slice profile due to Gz, z was scaled with λ 8:
$$\lambda = \frac{G_{slice}}{G_{slice} + G_z}$$
The integral in Equation (1) was solved by numerical integration with the following assumptions for each slice: linear field Δω(z,zi)=Δω0(zi)+Gz(zi)*(z-zi), φ0(zi)=const. and TR>>T1. Gz was obtained from the ΔB0-map.
MRI sequence: An adapted multi-echo gradient echo sequence (mGRE) was developed with two different sinc-Hanning-windowed pulses and variable polarity of Gslice (R2*-mapping: Bandwidth-Time-Product BWT=2.7 and pulse duration Tpulse=2ms, MWF-mapping: BWT=2; Tpulse=1ms).
Phantom R2*-mapping: A plastic cylinder (Ø=12cm; length=25cm) was filled with 110µmol/l Magnevist® doped agarose gel (5g/l) and scanned with positive and negative Gslice polarity (α=30°/90°; TE1/ΔTE/TR=5ms/5ms/3s; 32 monopolar echoes BW=500Hz/Px; resolution=1x1x4mm3; 25 slices). B1-map was derived from Bloch-Siegert shift9.
In-vivo R2*-mapping: Three subjects were scanned with the mGRE sequence with positive and negative Gslice (α=90°; TE1/ΔTE/TR=5ms/5ms/2.5s; 20 monopolar echoes BW=500Hz/Px; resolution=1x1x2mm3; 31 slices) and B1-mapping.
In-vivo MWF-mapping: One subject was scanned with the mGRE sequence (positive Gslice; α=90°; TE1/ΔTE/TR=2.37ms/2.2ms/2s; 32 bipolar echoes BW=500Hz/Px; resolution=1.1x1.1x4mm3; 25 slices; two averages; scan time 11min).
All measurements were performed on a clinical 3T scanner (Magnetom PRISMA, Siemens).
Analyses: For R2*-mapping F(zi,t) was calculated with and without B1+ and λ-correction. Then Equation (3) was minimized to fit R2*. For comparison, R2* was estimated from a conventional fit (F(zi,t)=1). MWF-maps were obtained by fitting once the measured data and once the data divided by F(zi,t) (regularized) to a multi-exponential model10. Cut-off for myelin water T2*my < 25ms11.
Results
Phantom: Figure 1 shows R2* as function of |Gz| from a conventional fit (F(zi,t)=1). Compared to α=30°, R2* for α=90° is different for +/- Gz and curves are flipped when Gslice polarity is changed. The normalized signal decay (Figure 1) for |Gz|=100µT/m reveals different signal decay for +/- Gz. Incorporating F(zi,t) resulted in a substantial, up to 5-fold, decrease of R2* compared to conventional fitting (Figure 2). Given this strong effect, the difference between using F(zi,t) with or without including B1+ and λ was smaller, but still visually observable.
In-vivo: Figure 3 shows R2*-maps for +/- Gslice as well as their absolute difference maps. While conventional fitted R2* show substantial differences, results with F(zi,t) are less influenced. The difference between the models in Figure 4 illustrates that B1+, λ-correction is most effective in areas with high B1+-deviations and strong Gz (e.g. frontal lobe).
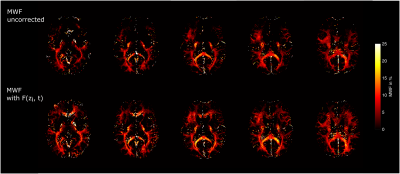
Uncorrected and corrected MWF-maps with F(zi,t) are illustrated in Figure 5. By using F(zi,t) it is possible to recover MWF values from the frontal lobe where the conventional fit fails.
Discussion and Conclusion
We have presented a comprehensive numerical model to
estimate the dephasing function F(zi,t) for an
arbitrary RF-pulse and slice selection gradient in the presence of Gz. We have demonstrated that the phase along the slice profile is the key
factor for modelling F(zi,t) and that the polarity of the slice selection gradient has to be taken into
account.
Furthermore, we showed that B1+ variations, especially
for high α, and strong Gz affect the slice profile and thus F(zi,t).
Besides R2*-mapping, myelin water imaging is an application
where our model yields strong improvements in areas affected by Gz (Figure 5).Acknowledgements
No acknowledgement found.References
1. Hirsch, N. M. & Preibisch, C. T2* mapping with background gradient correction using different excitation pulse shapes. AJNR. Am. J. Neuroradiol. 34, E65-8 (2013).
2. Hernando, D., Vigen, K. K., Shimakawa, A. & Reeder, S. B. R2*mapping in the presence of macroscopic B0field variations. Magn. Reson. Med. 68, 830–840 (2012).
3. Baudrexel, S. et al. Rapid single-scan T2*-mapping using exponential excitation pulses and image-based correction for linear background gradients. Magn. Reson. Med. 62, 263–268 (2009).
4. Fernndez-Seara, M. A. & Wehrli, F. W. Postprocessing technique to correct for background gradients in image-based R2* measurements. Magn. Reson. Med. 44, 358–366 (2000).
5. Yablonskiy, D. A. Quantitation of intrinsic magnetic susceptibility-related effects in a tissue matrix. Phantom study. Magn. Reson. Med. 39, 417–428 (1998).
6. Preibisch, C., Volz, S., Anti, S. & Deichmann, R. Exponential excitation pulses for improved water content mapping in the presence of background gradients. Magn. Reson. Med. 60, 908–916 (2008).
7. Aigner, C. S., Clason, C., Rund, A. & Stollberger, R. Efficient high-resolution RF pulse design applied to simultaneous multi-slice excitation. J. Magn. Reson. 263, 33–44 (2016).
8. Reichenbach, J. R. et al. Theory and application of static field inhomogeneity effects in gradient-echo imaging. J. Magn. Reson. Imaging 7, 266–279 (1997).
9. Sacolick, L. I., Wiesinger, F., Hancu, I. & Vogel, M. W. B1 mapping by Bloch-Siegert shift. Magn. Reson. Med. 63, 1315–1322 (2010).
10. Whittall, K. P. & MacKay, A. L. Quantitative interpretation of NMR relaxation data. J. Magn. Reson. 84, 134–152 (1989).
11. Lenz, C., Klarhöfer, M. & Scheffler, K. Feasibility of in vivo myelin water imaging using 3D multigradient-echo pulse sequences. Magn. Reson. Med. 68, 523–528 (2012).
Figures