4855
k-space deep learning for MR herringbone artifact correction1KAIST, Daejeon, Korea, Republic of
Synopsis
Herringbone artifact is caused by power fluctuation of MR equipment or unstable shielding. Herringbone artifact image is difficult to analyze because it scatters on whole image region of single or multiple slices. There is a study for MR artifact correction which can be represented as sparse outliers on k-space. This method exploits the duality between the low-rankness of Hankel matrix in k-space and the sparsity in the image domain. However, this method has high computational complexity, and consumes much time. In this research, we suggest the new effective and fast MR artifact correction method using deep learning.
Introduction
There are some MR artifacts due to hardware or software problems. Herringbone artifact, which is also called criss-cross artifact, is caused by power fluctuation of MR equipment or unstable shielding.[1] Herringbone artifact image is difficult to analyze because it scatters on whole image region of single or multiple slices. There is a study for MR artifact correction which can be represented as sparse outliers on k-space.[2] Most MR artifacts shows some sparse outliers on frequency domain, and herringbone artifact shows point-like spike noises on k-space, especially. Therefore, they exploits the duality between the low-rankness of Hankel matrix in k-space and the sparsity in the image domain to reduce artifact. However, this method has high computational complexity, so it consumes much time.
Nowadays, many researchers try to apply deep neural network for MR study. Deep convolutional framelet theory[3] reveals that the deep neural network can be explained by an extension of classical signal processing theory. Inspired by this theory, we expected the deep neural network with encoder-decoder architecture also works for k-space. In this research, we suggest the new effective and fast MR artifact correction method using k-space deep learning.
Methods
7T T1 weighted data are acquired from Phillips whole body MR scanner(Phillips Achiva system) from 9 subjects with 32 channel head coils. Acquisition parameters were as follows: TR/TE = 831/5 $$$ms$$$, 63 slices with 0.75 $$$mm$$$ slice thickness, FOV of 220x220$$$mm^2$$$ , and 288x288 matrix size with 3/4 partial fourier sampling. Fig.1 shows the flow and network structure of this work. The real in-vivo data are used as labels, and random impulse noise is added on the k-space measurement to make noisy input data. If the intensity of the impulse is similar to the k-space signal, it is less sparse than the high intensity case. To observe whether k-space learning method removes only impulse noise well for different level of noise, we also used another noise data corresponding to one third of the impulse intensity level of training data on the inference step. Also the weighting function is applied to the input k-space to make k-space more sparse, and the result of neural network is unweighted by using same weighting function. Based on deep convolutional framelets theory[3] that explains a relationship between neural network with the encoder-decoder architecture and hankel matrix decomposition, we suggested a deep learning method on k-space. The training network is worked on k-space domain, but loss is calculated in the image domain. The detailed network structure is shown in (b). For basic module, there are 2 sets of 3x3 convolution with batch normalization and ReLU layer(red arrow). This module is repeated 5 times with 2x2 pooling and unpooling layers. Also, intermediate images are concatenated on decoding parts.(blue arrow) Because k-space consists of complex values, the layer of changing from complex to real value and the layer of changing from real to complex value are placed at the beginning. Among 9 subjects, we used 7 subjects for training, 1 subject data for validation, and 1 subject data for test. The network was implemented using MatConvNet toolbox(ver.24) in MATLAB 2015a environment (Mathwork, Natick). We used a GTX 1080 Ti graphic processor and i7-4770 CPU (3.40GHz).Results
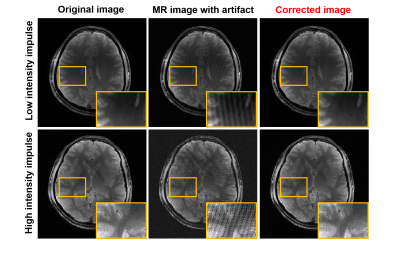
Fig.2 shows corrected image and corresponding k-space. The neural network effectively removes impulse noises on k-space. Also, herringbone artifacts on image domain are corrected clearly.(fig.3) For both cases of impulse noise intensity level data, the proposed network shows clear images without any noise. That is, the network is well trained to remove impulse noise on k-space even for the low intensity impulse. It tooks only 0.0155s to 32 multi-coil and 40 slices of test subject data while the conventional method[1] could not correct artifact due to the large data size.Conclusion
This work shows the novel MR artifact correction method using deep learning method. The proposed method shows good performance to remove impulse noises on k-space and noise on image domain. We expect that this method becomes to apply other MR artifact research.Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea(NRF) funded by the Ministry of Science and ICT (NRF-2016R1A2B3008104)
References
1. A.Stadler et al, European radiology, 17.5 (2007) 2. K.H.Jin et al, Magn Reson Med 78 (2017) 3. J.C.Ye et al, Siam J Imaging Sci 11.2 (2018)