4818
Brain tissue segmentation in fetal MRI using convolutional neural networks with simulated intensity inhomogeneities1Image Sciences Institute, Utrecht University, Utrecht, The Netherlands, Utrecht, Netherlands, 2Department of Neonatology, Wilhelmina Children’s Hospital, University Medical Center Utrecht, The Netherlands, Utrecht, Netherlands, 3Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, The Netherlands, Utrecht, Netherlands
Synopsis
Automatic brain tissue segmentation in fetal MRI is a challenging task due to artifacts such as intensity inhomogeneity, caused in particular by spontaneous fetal movements during the scan. Unlike methods that estimate the bias field to remove intensity inhomogeneity as a preprocessing step in
Introduction
MRI is commonly used for monitoring and detecting abnormalities in fetal development, particularly in fetal brain development [1]. Segmentation of different tissue classes in the fetal brain is a prerequisite for early detection of abnormalities, but manual segmentation is an extremely time-consuming task and requires a high level of expertise. Automatic segmentation is challenging as well due to the low of contrast between different tissue classes, motion artifacts and image artifacts such as intensity inhomogeneity (see Figure 1). Fetal MRI is acquired using phased-array coils on the maternal body and the signal intensity can drop quickly with fetal movements during the scanning. Training supervised segmentation methods such as convolutional neural networks that are robust to intensity inhomogeneity is often limited by the number of available manual reference segmentations of slices with this type of artifacts, given the cumbersome process of manual annotation of slices with artifacts. We therefore propose to synthesize slices with intensity inhomogeneities from artifact free slices with corresponding reference segmentations, which we refer to as intensity inhomogeneity augmentation (IIA).Method
T2-weighted MR scans of 12 fetuses (22.9-34.6 weeks postmenstrual age) were included in this study. Images were acquired on a Philips Achieva 3T scanner using a turbo fast spin-echo sequence. The acquired voxel size was 1.25×1.25×2.50 $$$mm^3$$$ and the reconstructed voxel size was 0.70×0.70 ×1.250 $$$mm^3$$$. Fetal MRI has a large field of view which visualizes the maternal body as well as the entire fetal body. The intracranial volumes (ICV) were therefore first automatically segmented to mask the fetal brains [2]. The masked images were segmented into four tissue types using a U-net [3] (see Figure 2) trained with 2D fetal MRI slices. Each pixel in the image was classified as either white matter (WM), gray matter (GM), extracerebral or ventricular cerebrospinal fluid (CSF), basal ganglia or thalamus or brainstem (BGT-BS), or background. The network was trained by maximizing the Dice coefficient between network output and manual segmentation using stochastic gradient descent. To make the segmentation network robust to intensity inhomogeneities, we trained the network with both artifact free slices (70%), and with slices showing simulated intensity inhomogeneity (30%). Intensity inhomogeneity was simulated by applying a combination of linear gradients with random offsets and orientations to artifact free slice I: $$Z = I\times((X+x_0)^{2}+(Y+y_0)^{2}),$$ where X and Y are 2D matrixes with integer values from zero to the size of the image in x and y direction, respectively. The offsets $$$x_0$$$ and $$$y_0$$$ control the balance between the x and y components and were randomly chosen from different ranges ($$$x_0$$$: [43, 187]; $$$y_0$$$: [-371, 170]). The optimal ranges were found with a random hyperparameter search. Additionally, the gradient patterns were randomly rotated to mimic intensity inhomogeneity in various directions. These two random components result in inhomogeneity patterns that allow the network to become invariant to the location and orientation of regions with low and decreasing contrast. The intensities in both the original slices as well as the slices with simulated intensity inhomogeneity ($$$Z$$$) were normalized to the range [0, 1024] before feeding them to the network. Moreover, other data augmentation techniques such as random flipping and rotation were applied to further increase the variation in the training data.
Experiments and Results
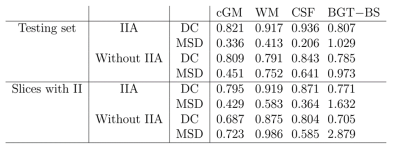
The automatic segmentation was evaluated in 2D by means of Dice coefficient (DC) and the mean surface distance (MSD) between manual and automatic segmentation. The network was trained with 6 fetal coronal scans and was tested with the remaining 6 scans. Slices with intensity inhomogeneity were removed from the training set, but not from the test set. Comparing the network trained with IIA with a network trained without IIA showed that the segmentation performance improved for all tissues, in particular for WM (Table 1). The performance improvement was especially notable in slices with intensity inhomogeneity (Figure 4), of which only simulated samples were part of the training set.Conclusion
Intensity inhomogeneity is a frequent artifact in MRI and especially affects imaging if movements of the subject cannot be effectively reduced, such as in fetal MRI. We demonstrated that supervised image processing methods, such as convolutional neural networks, can become more robust to these artifacts by training them with data having simulated intensity inhomogeneities. This can potentially replace or complement preprocessing steps, such as bias field corrections. Simulating artifacts instead of collecting compromised data is beneficial as manual reference annotations can be obtained more easily and with higher accuracy for artifact free data. In future work, more complex inhomogeneity patterns could be investigated.
Acknowledgements
This study was financially supported by the Research Program Specialized Nutrition of the Utrecht Center for Food and Health, through a subsidy from the Dutch Ministry of Economic Affairs, the Utrecht Province and the Municipality of Utrecht.References
[1] D. M. Twickler, T. Reichel, D. D. McIntire, K. P. Magee, R. M. Ramus, Fetal central nervous system ventricle and cisterna magna measurements by magnetic resonance imaging, American journal of obstetrics and gynecology 187 (4) (2002) 927–931.
[2] N. Khalili, P. Moeskops, N. Claessens, S. Scherpenzeel, E. Turk, R. de Heus, M. Benders, M. Viergever, J. Pluim, I. Iˇsgum, Automatic segmentation of the intracranial volume in fetal MR images, in: Fetal, Infant and Ophthalmic Medical Image Analysis, 2017, pp. 42–51. [3] O. Ronneberger, P. Fischer, T. Brox, U-net: Convolutional networks for biomedical image segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, 2015, pp. 234–241.
Figures