4788
A divide-and-conquer strategy to overcome memory limitations of current GPUs for high resolution MRI reconstruction via a domain transform deep learning method1Medical Physics, University of Wisconsin-madison, Madison, WI, United States, 2Radiology, University of Wisconsin-madison, Madison, WI, United States
Synopsis
Direct learning of a domain transform to reconstruct images with flexible data acquisition schemes represents a step to achieve intelligence in image reconstruction. However, a technical challenge that is encountered with the domain transform type of learning strategy is that current network architectures and training strategies are GPU memory hungry. As a result, given the currently available GPUs with memory on the order of 24 GB, it is very difficult to achieve high resolution (beyond 128x128) MRI reconstruction. The main purpose of this paper is to present a divide-and-conquer strategy to reconstruct high resolution (better than 256x256) MRI images via domain transform learning while staying within the current GPU memory restrictions.
Introduction
Several deep learning reconstruction strategies have been proposed to learn a prior model to regularize iterative reconstruction1, to learn the strength and intrinsic parameters in an expert field prior model2, to learn a denoising convolutional neural network (CNN) to cascade with the data fidelity solver in reconstruction3, and more recently, to directly learn to perform a domain transform from k-space to image followed by a CNN to correct the resulting image4. The potential advantages provided by the direct learning of a domain transform is to enable one to reconstruct images with flexible data acquisition schemes, a needed step to achieve truly intelligent image reconstruction. However, one technical challenge that is encountered with the domain transform type of learning strategy is that the current network architectures and training strategies are GPU memory hungry. As a result, given the currently commercially available GPUs with memory on the order of 24 GB, it is very difficult, if not impossible to achieve high resolution MRI reconstruction with a domain transform learning strategy. The main purpose of this paper is to present a divide-and-conquer strategy to reconstruct high resolution MRI images via domain transform learning while staying within the current GPU memory restrictions.Method
The central idea of our strategy is to train the domain transform learning scheme to reconstruct smaller patches of the image, instead of the whole image in one step. Suppose the dimension of the input k-space data is $$$2n^2$$$ and the target reconstruction patch is $$$m^2$$$. We design a neural network architecture with $$$s=n^2/m^2$$$ branches. Each of these branches is independently trained to perform the patch reconstruction. As an example (Fig.1), 16 branches were trained in our architecture to achieve the reconstruction of 16 patches with dimension of 64x64. These patches are stitched seamlessly to obtain the final 256x256 image matrix.
In our proposed network architecture, there is a single fully-connected layer to transform data from k-space to image space. Following the fully-connected layer is a shallow U-Net-like network structure to perform image correction such that the reconstructed image patches are matched with training labels in terms of minimal least square error loss. This architecture is different from AUTOMAP in two aspects: our architecture consists of a single fully-connected layer, while there are two in AUTOMAP; the second difference is that we used a U-Net-like CNN to clean up the image as opposed to the 3-layer auto-encoder used in AUTOMAP. In each branch, the single fully-connected layer is followed by a hyperbolic tangent activation and ReLu activation is used in the rest of the layers. The output layer is a 1x1 convolution transpose layer with a linear activation. A concatenation operation from the output of the third layer to the sixth layer was utilized to help preserve spatial resolution (Fig.1).
The network was first pre-trained using 10,000 images from an anonymized MR dataset with a factor of four augmentation to prevent overfitting. A separate set of 1,000 images were reserved as the test set. Phase modulation was embedded into the images to simulate reconstruction of MR complex images. In order to accommodate the MR acquisition, the network was then fine-tuned using experimental data acquired from an MRI scanner. Finally, separate k-space samples were used as the final test set.
In both pre-training and fine-tuning, the ADAM optimizer was used with a momentum of 0.9. The learning rate was set to 2e-4 for 50 epochs and then lowered to 2e-5 for an additional 50 epochs, with a minibatch size of 50. The network can be trained on a single NVIDIA GTX 1080ti with 11 GB of memory.
Results
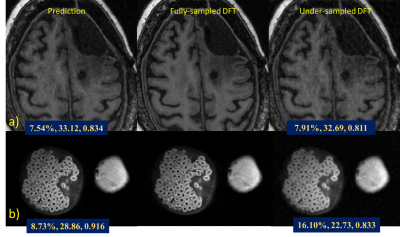
The results of the fully-sampled k-space reconstruction (Fig.2a) demonstrate that the proposed network and divide-and-conquer strategy can accurately reconstruct the phase patterns in the real and imaginary components of the image and thus output the correct final magnitude image. The visual performance shows no significant difference between the network prediction and reference image. The fine-tuned network incorporated non-ideal factors in raw scanner acquisitions and could help mitigate spatial resolution loss (Fig.2b). Factor of three under-sampled k-space reconstruction results are shown in Fig.3a and Fig.3b. Quantitative image quality assessment metrics of rRMSE, PSNR, SSIM were used to quantify the reconstruction quality.Conclusion
The proposed divide-and-conquer neural network architecture and corresponding training strategy enable one to reconstruct high spatial resolution MRI images via a domain transform learning strategy. The proposed method can be equally applicable to other domain transform learning strategies such as AUTOMAP.Acknowledgements
No acknowledgement found.References
1. Wang S, Su Z, Ying L, Peng X, Zhu S, Liang F, Feng D, Liang D. Accelerating magnetic resonance imaging via deep learning. InBiomedical Imaging (ISBI), 2016 IEEE 13th International Symposium on 2016 Apr 13 (pp. 514-517). IEEE.
2. Hammernik K, Klatzer T, Kobler E, Recht MP, Sodickson DK, Pock T, Knoll F. Learning a variational network for reconstruction of accelerated MRI data. Magnetic resonance in medicine. 2018 Jun;79(6):3055-71
3. Schlemper J, Caballero J, Hajnal JV, Price AN, Rueckert D. A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE transactions on Medical Imaging. 2018 Feb;37(2):491-503.
4. Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature. 2018;555(7697):487-492. doi:10.1038/nature25988.
Figures

Proposed network architecture with 16 branches. denotes the U-NET-like convolutional neural network.