4730
Automated organ segmentation of liver and spleen in whole-body T1-weighted MR images: Transfer learning between epidemiological cohort studies1School of Biomedical Engineering & Imaging Sciences, King's College London, London, United Kingdom, 2Department of Radiology, University Hospital Tübingen, Tübingen, Germany, 3Institute of Signal Processing and System Theory, University of Stuttgart, Stuttgart, Germany
Synopsis
Automated segmentation of organs and anatomical structures is a prerequisite for efficient analysis of MR data in large cohort studies with thousands of participants. The feasibility of deep learning approaches has been shown to provide good solutions. Since all these methods are based on supervised learning, labeled ground truth data is required which can be time- and cost-intensive to generate. This work examines the feasibility of transfer learning between similar epidemiological cohort studies to derive possibilities in reuse of labeled training data.
Introduction
Automated analysis of imaging data plays an important role for rapidly increasing numbers of large cohort studies and information content per study. Especially in large epidemiological imaging studies with thousands of scanned individuals, a visual and manual image processing is no longer feasible. In order to target any automated analysis, one crucial step in the evaluation of medical image content is the recognition and segmentation of organs. Several methods applying automated organ segmentation to MR images have been proposed so far, including methods relying on explicit models1, general correspondence2, machine learning such as Random Forests3 as well as more recently deep learning4 (DL) approaches including convolutional neural networks (CNNs)5. Amongst these methods, DL6,7 demonstrates excellent performance. All schemes share the goal to infer class labels by generalizing well from labeled training data. However, the creation of labeled training data can be time-consuming and cost-intensive, because it needs to be performed by trained experts. Reuse of already labeled training data is wherever suitable therefore of high interest.
In this study, we examine the feasibility of a transfer learning between two similar epidemiological MR imaging databases: Cooperative Health Research in the Region of Augsburg (KORA)8 and German National Cohort (NAKO)9. We utilize our previously proposed DCNet10,11 to automatically segment liver and spleen in both databases and investigate the capabilities of a transfer learning.
Material and Methods
In the context of the KORA MR study, coronal whole-body T1-weighted dual-echo gradient echo images were acquired for 397 subjects on a 3T MRI. The imaging parameters are: 1.7mm isotropic resolution with matrix size=288x288x160, TEs=1.26/2.52ms, TR=4.06ms, flip angle=9°, bandwidth=755Hz/px. For the NAKO MR study, transversal whole-body T1-weighted dual-echo gradient echo images were acquired for 200 subjects on a 3T MRI with imaging parameters: 1.4x1.4x3 mm, TEs=1.23/2.46ms, TR=4.36ms, flip angle=9°, bandwidth=680Hz/px. Subsets of 173/97 for KORA/NAKO were randomly selected and liver and spleen were manually labeled by experienced radiologists.
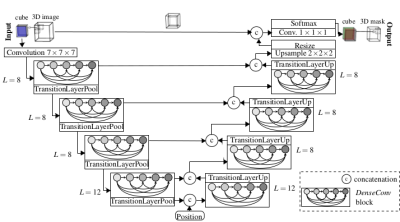
The proposed DCNet10,11 (Fig.1) combines the concepts of UNet12 for pixel-wise localization, VNet7 for volumetric medical image segmentation, ResNet13 to deal with vanishing gradients and degradation problems and DenseNet14 to enable a deep supervision. For the task of semantic segmentation, additional positional information is fed to the network serving as a-priori knowledge of relative organ positioning. This is achieved by concatenating global scanner coordinates of the respective VOI to the feature maps after the encoding branch.
Input images are cropped to 3D patches of size 32x32x32 with 2-channel input (fat and water image) enabling better delineation of organs. An RMSprop trains the network with Jaccard distance loss, batch size=48 and over 50 epochs over 4 runs to investigate stability. Different training cases were conducted to evaluate the influence of transfer learning on the classification result (Tab.1).
Results and Discussion
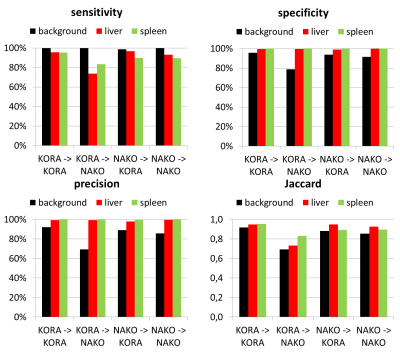
Overall, a very good agreement of automated organ segmentation of liver and spleen was observed compared to manual segmentation as shown exemplary in Fig.2. The smaller sized spleen can be well delineated simultaneously to the larger sized liver, although their similar head-feet position. Best results were obtained if the networks were trained and evaluated on the same database (case 1 and 4). Despite the anisotropic resolution in axial direction and the stitched multi-bed FOVs of the NAKO dataset, the network’s prediction (KORA->NAKO) is still of high accuracy indicating generalizability of the proposed method. However, backward prediction (NAKO->KORA) is losing specificity for anisotropic NAKO to isotropic KORA prediction. The positional input encoding helps to reduce false outliers especially in the transfer learning context (Fig.3). High quantitative metrics (Fig.4) are obtained for all classes in the cases 1 and 4, whilst these values are slightly reduced in the transfer learning cases 2 and 3. Background, respectively outliers, is increased in the transfer learning cases indicated by reduced precision and Jaccard.
Our study has limitations. The obtained results are specific for the imaging setup and MR sequence design of the study at hand. The segmented organs in our study (liver and spleen) are rather large structures in the upper abdomen and may thus be easier to segment compared to more complicated anatomic structures. Thus, generalizability to other imaging sequences and organs will be investigated in future studies.
Conclusion
In this study we investigated the possibility of automated segmentation of two abdominal organs in T1-weighted MR images using our proposed deep neural network (DCNet). A transfer learning is feasible and labeled ground truth can be reused if imaging databases share commonalities (imaging sequence, contrast weighting). Accurate automated segmentation of organs in epidemiological cohort studies with reduced effort for generating labeled training data seems feasible via the proposed deep learning approach.Acknowledgements
No acknowledgement found.References
1. Heimann T, Meinzer H-P. Statistical shape models for 3D medical image segmentation: a review. Medical image analysis 2009;13(4):543-563.2. Iglesias JE, Sabuncu MR. Multi-atlas segmentation of biomedical images: a survey. Medical image analysis 2015;24(1):205-219.
3. Criminisi A, Shotton J, Konukoglu E, others. Decision forests: A unified framework for classification, regression, density estimation, manifold learning and semi-supervised learning. Foundations and Trends in Computer Graphics and Vision 2012;7(2--3):81-227.
4. Shen D, Wu G, Suk H-I. Deep learning in medical image analysis. Annual review of biomedical engineering 2017;19:221-248.
5. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, S\'a n, Clara I. A survey on deep learning in medical image analysis. Medical image analysis 2017;42:60-88.
6. Lavdas I, Glocker B, Kamnitsas K, Rueckert D, Mair H, Sandhu A, Taylor SA, Aboagye EO, Rockall AG. Fully automatic, multiorgan segmentation in normal whole body magnetic resonance imaging (MRI), using classification forests (CFs), convolutional neural networks (CNNs), and a multi-atlas (MA) approach. Medical Physics 2017;44(10):5210-5220.
7. Milletari F, Navab N, Ahmadi S-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. ArXiv e-prints 2016.
8. Holle R, Happich M, Löwel H, Wichmann HE, study group MKORA, others. KORA-a research platform for population based health research. Das Gesundheitswesen 2005;67(S 01):19-25.
9. Bamberg F, Kauczor HU, Weckbach S, Schlett CL, Forsting M, Ladd SC, Greiser KH, Weber MA, Schulz-Menger J, Niendorf T, Pischon T, Caspers S, Amunts K, Berger K, Bulow R, Hosten N, Hegenscheid K, Kroncke T, Linseisen J, Gunther M, Hirsch JG, Kohn A, Hendel T, Wichmann HE, Schmidt B, Jockel KH, Hoffmann W, Kaaks R, Reiser MF, Volzke H. Whole-Body MR Imaging in the German National Cohort: Rationale, Design, and Technical Background. Radiology 2015;277(1):206-220.
10. Küstner T, Fischer M, Müller S, Gutmann D, Nikolaou K, Bamberg F, Yang B, Schick F, Gatidis S. Automated segmentation of abdominal organs in T1-weighted MR images using a deep learning approach: application on a large epidemiological MR study. Proceedings of the International Society for Magnetic Resonance in Medicine (ISMRM); 2018.
11. Küstner T, Müller S, Fischer M, Weiβ J, Nikolaou K, Bamberg F, Yang B, Schick F, Gatidis S. Semantic Organ Segmentation in 3D Whole-Body MR Images. 2018 25th IEEE International Conference on Image Processing (ICIP); 7-10 Oct. 2018 2018. p 3498-3502.
12. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention; 2015. p 234-241.
13. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016. p 770-778.
14. Huang G, Liu Z, Weinberger KQ, van der Maaten L. Densely connected convolutional networks. arXiv preprint arXiv:160806993 2016.
Figures