4728
The deep learning lesion segmentation method nicMSlesions only needs one manually delineated subject to outperform commonly used unsupervised methods1Department of Radiology and Nuclear Medicine, MS Center Amsterdam, Amsterdam Neuroscience, Amsterdam UMC - location VUmc, Amsterdam, Netherlands, 2Institutes of Neurology and Healthcare Engineering UCL, London, United Kingdom
Synopsis
Automatic lesion segmentation is important for measurements of atrophy and lesion load in subjects with multiple sclerosis (MS). Although supervised methods perform overall better than unsupervised methods, they are not widely used since they are more labor-intensive due to the need for great amounts of manual input. Our research showed increased performance of supervised methods over unsupervised methods. In addition, when using a deep learning based supervised method, training on only one subject already outperformed the commonly used unsupervised methods. We therefore recommend using deep learning lesion segmentation methods in MS research.
Background
Multiple sclerosis (MS) is an autoimmune disorder of the central nervous system, characterized by neurodegeneration and demyelination. To enable both atrophy and lesion load measurements in subjects with MS, accurate lesion segmentation is necessary. Over the last decade, several (semi-)automatic lesion segmentation methods have been developed, which can be divided into two groups: supervised methods, which require manual delineation to train the method properly, and unsupervised methods, which do not require any training. Unsupervised methods are less labor-intensive, but show overall poor agreement with manual delineation.1Aims
Therefore, the aim of this research was twofold: first, we investigated the volumetric and spatial agreement of two supervised and two unsupervised automated lesion segmentation methods. Second, we assessed whether input from only one subject’s manual delineation in the deep learning based supervised method already improved the volumetric and spatial agreement over unsupervised methods.Methods
A total of fourteen subjects with RRMS
were scanned between December 2016 and June 2017 on a 3T whole-body MR scanner
(GE Discovery MR750) with an 8-channel phased-array head coil. The protocol
included a 3D T1-weighted fast spoiled gradient echo sequence (FSPGR with
TR/TE/TI = 8.2/3.2/450 ms and resolution 1.0x1.0x1.0 mm) and a 3D T2-weighted
fluid attenuated inversion recovery sequence (FLAIR with TR/TE/TI =
8000/130/2338 ms at resolution 1.0x1.0x1.2 mm). An expert rater (experience
>10 years) manually delineated the lesions on FLAIR images. Next, a total of
four automated lesion segmentation methods were tested in comparison to manual
segmentation, all based on different underlying algorithms. We tested two
unsupervised methods, i.e. Lesion-Topology preserving Anatomical Segmentation (LesionTOADS)2 and
Lesion Segmentation Toolbox with Lesion Prediction Algorithm (LST)3 and two supervised methods, i.e. FMRIB Software Library’s Brain
Intensity AbNormality Classification Algorithm (FSL BIANCA)4 and
Valverde’s nicMSlesions5. For the
two supervised methods, we used input from all fourteen manually delineated subjects
and used leave-one-out cross-validation.
For LST LPA, the probability threshold
was set on 0.55 as reported previously1. BIANCA
was optimized on our dataset (all and equal number of lesion and non-lesion points
in the training set, any location of the non-lesion training points, a 3D patch
with patch size 5, spatial weighting of 2, and threshold 0.99). For
nicMSlesions, default parameters were used with threshold 0.5. No optimization
was needed for LesionTOADS.
For our second aim, we further tested
nicMSlesions with input from only one manually delineated subject for its
performance on the other thirteen subjects. We looked at volumetric and spatial
agreement of the various methods compared to manual, using repeated measures
ANOVA with, when appropriate, post-hoc Wilcoxon Signed Ranks testing. Results
were considered significant upon p
< 0.05.
Results
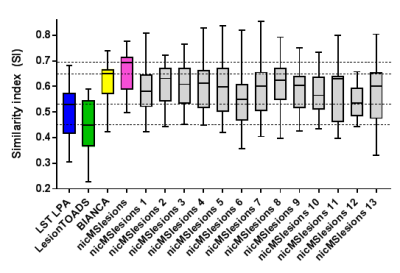
An example of the performance of the manual and automatic lesion segmentation methods is visualized in Figure 1 (note: here, the supervised methods are trained on fourteen subjects with leave-one-out cross-validation). The automated segmentation method used significantly affected both volumetric agreement (F(4,52) = 25.650, p < 0.001) and spatial agreement (F(3,39) = 27.954, p < 0.001) (Table 1). Post-hoc testing showed that manual volumes differed significantly from those of LST LPA, LesionTOADS and nicMSlesions, but not from those of BIANCA (Figure 2).For the single-subject training of nicMSlesions, one subject failed the training and was therefore not included in any of the further analyses. The single-subject (1 to 13) that was used for training significantly affected the volumetric agreement (F(13,156) = 25.465, p < 0.001), but not the spatial agreement (F(12,144) = 1.497, p = 0.132) (Table 2). All single-subject trained nicMSlesions variants had greater spatial agreement with manual than the unsupervised lesion segmentation methods (Figure 3).
Discussion and conclusion
The two supervised methods showed better volumetric and spatial agreement to manual than the unsupervised methods, with BIANCA showing the best volumetric and nicMSlesions showing the best spatial agreement. Since the settings of nicMSlesions were set to default, it is possible that better volumetric agreement can be obtained upon optimization of the method.Furthermore, our results show that manual lesion segmentation input from even one single subject is sufficient to train nicMSlesions with its default parameters in such a way that it outperforms the unsupervised methods LST LPA and LesionTOADS. Although training on multiple subjects shows even better volumetric and spatial agreement, studies without great amounts of manual delineations can use nicMSlesions with only one subject’s input and improve their automatic lesion segmentation over the commonly used unsupervised methods.
Results should be confirmed in multi-vendor images and in subjects with different MS phenotypes.
Acknowledgements
This work was supported by the Dutch MS Research Foundation (grant number 14-876).
References
- de Sitter A, Steenwijk MD, Ruet A, Versteeg A, Liu Y, van Schijndel RA, et al. Performance of five research-domain automated WM lesion segmentation methods in a multi-center MS study. Neuroimage. 2017;163:106-14.
- Shiee N, Bazin PL, Ozturk A, Reich DS, Calabresi PA, Pham DL. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. Neuroimage. 2010;49(2):1524-35.
- Schmidt P. Bayesian Inference for Structured Additive Regression Models for Large-scale Problems with Applications to Medical Imaging: LMU München; 2017.
- Griffanti L, Zamboni G, Khan A, Li L, Bonifacio G, Sundaresan V, et al. BIANCA (Brain Intensity AbNormality Classification Algorithm): A new tool for automated segmentation of white matter hyperintensities. Neuroimage. 2016;141:191-205.
- Valverde S, Cabezas M, Roura E, Gonzalez-Villa S, Pareto D, Vilanova JC, et al. Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. Neuroimage. 2017;155:159-68.
Figures