4707
Development of a deep learning method for phase unwrapping MR images1Department of Electrical and Electronic Engineering, Yonsei University, Seoul, Korea, Republic of, 2MR Clinical research and Development, GE Healthcare, Seoul, Korea, Republic of, 3Seoul St.Mary's Hospital, The Catholic University of korea, Seoul, Korea, Republic of, 4Department of Radiology, Medical College of Wisconsin, Milwaukee, WI, United States
Synopsis
MRI phase images are increasingly used for susceptibility mapping and distortion correction in function and diffusion MRI. However, acquired values of phase maps are wrapped between [-π π ] and require an additional phase unwrapping process. Here we developed a novel deep learning method that can learn the transformation between the wrapped phase images and the corresponding unwrapped phase images. The method was tested for numerical simulations and on actual MR images.
Introduction
Phase images in MRI are used in various applications1-4. Typical phase images require dedicated phase unwrapping algorithms which have trade-off between high computation times and accuracy5. Thus, deep learning (DL) methods which are both time efficient and accurate may have advantages. Conventionally used deep learning architectures6 used in medical imaging are difficult to apply directly. Architectures such as UNET7 are based on convolutional layers and have difficulty in learning the global features of the image due to receptive field constraints. In addition, loss functions such as mean squared error (MSE) is insufficient for the problem as the solution is not unique, i.e., if Y is one solution (unwrapped) for arbitrary integer n, Y+2nπ may also be solutions in phase unwrapping. We developed a DL method that can resolve the aforementioned problems. Specifically, we utilized recurrent modules that can efficiently learn global features and an appropriate loss function based on the contrast of the error. Numerical simulations and in vivo studies with comparison to 3D PRELUDE8,9, which is often regarded as the gold standard, is performed.
Methods
[Network architecture]
Figure 1 (a) shows the proposed deep neural network design. The key feature of the architecture is the bidirectional RNN (recurrent NN) module10. This module reads through each pixel following the four paths (Left to right, right to left, down to up, up to down), enabling global spatial features to be learned. Conventional UNET was also trained and compared.
[Loss function]
Loss function was composed of two loss functions, namely total variation loss and variance of error loss. Total variation loss was $$$L_{TV} (\widehat{Y},Y)= E[|∇_x (\widehat{Y}-Y)|+|∇_y (\widehat{Y}-Y)|]$$$ and the variance of error loss was $$$L_V (\widehat{Y},Y)= E[(\widehat{Y}-Y)^2 ]-(E[\widehat{Y}-Y])^2,$$$ where E[X] denotes the mean value of the image X. The final loss function was weighted by $$L(\widehat{Y},Y)=0.1*L_{TV} (\widehat{Y},Y)+ L_V (\widehat{Y},Y).$$ For in-vivo dataset, to reflect the fact that the phase SNR is inversely proportional to the magnitude intensity, the losses were further modified respectively as: $$ L_{TV} (\widehat{Y},Y)= (|∇_x M*(\widehat{Y}-Y)|_F+|∇_y M*(\widehat{Y}-Y)|_F)/∑M $$,
$$ L_{V} (\widehat{Y},Y)= E[M*(\widehat{Y}-Y)^2]/E[M]-(E[M*(\widehat{Y}-Y)]/E[M])^2,$$
where M is the magnitude image and * the element-wise multiplication.
[Dataset]
1. Simulation:
For numerical simulation, images w generated based on Eq.1 with randomly generated variables σa, σb, σc, M, xc, yc.
$$I(x,y)= σ_a x+σ_b x+σ_c+\sum_{n=1}^Me^{-{\frac{(x-x_c)^2}{2σ_x}+\frac{(y-y_c)^2}{2σ_y}}} [Eq.1]$$
The wrapped phase was synthetically generated by $$$\phi(x,y)= \angle{e^{jI(x,y)}}.$$$ An exemple dataset pair is shown in Fig.1 (b).
2. In-vivo
The paired dataset was generated by processing the PRELUDE on actual multi-echo GRE images. Dataset from ten subjects were used for training the network. An example dataset is shown in Fig.1 (c). mGRE images (8-echoes) from one subjects not included in the training were used for the testing.
Results
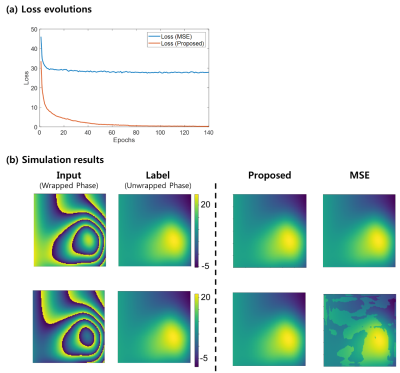
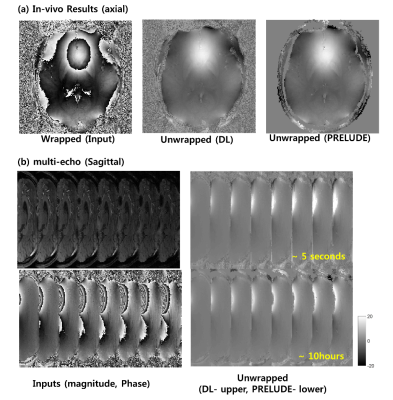
Figure 2 shows the performance of the proposed method over UNET architecture. Using U-Net the result was successful for local phase wraps (a) but fails on global phase wraps (b), whereas the proposed method was successful for both situations. Figure 3 shows the performance of the proposed loss function over MSE. The loss of the proposed loss function decreased to zero, while the MSE loss was limited at some level (a). Figure 3 (b) shows that the resultant images with the proposed method were successful compared to the MSE loss. Figure 4 shows an axial image (a), sagittal image (b). Note that DL method was processed slice by slice and concatenated on the slice direction, whereas the PRELUDE method was processed 3-dimensionally echo by echo. Comparative results are given while the reconstruction time was very different.Discussion and Conclusion
In this study, we developed and demonstrated the neural network and the loss function for training the phase unwrapping process. The proposed method was successful at learning the phase unwrapping process for both the numerically simulated images and in-vivo images. Moreover, the DL method was time efficient compared to the PRELUDE (5 seconds vs 10 hours). However, there are some limitations. First, for in-vivo training, the dataset was relatively small (10 subjects) and the label images contained some errors from PRELUDE process. More data and thorough data preparation will be explored to improve the results. Moreover, the current network is 2D based process and does not yet exploit the information from the slice directions. Further improvement of the network to learn 3-dimensional phase unwrapping will be explored.Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea(NRF) funded by the Ministry of Science, ICT and future Planning (NRF-2016R1A2B3016273).
The research was supported by Graduate Student Scholarship Program funded by Hyundai Motor Chung Mong-Koo Foundation.
References
1. Shmueli, Karin, et al. “Magnetic susceptibility mapping of brain tissue in vivo using MRI phase data.” Magnetic resonance in medicine 62.6 (2009): 1510-1522
2. Reichenbach, J.R., et al. “Quantitative susceptibility mapping: concepts and applications.” Clinical neuroradiology 25.2 (2015): 225-230.
3. Kelin, Tilmann A., Markus Ullsperger, and Gerhard Jocham. “Learning relative values in the striatum induces violations of normative decision making.” Nature Communications 8 (2017).
4. Shen, Kaikai, et al. “A spatio-temporal atlas of neonatal diffusion MRI based on kernel ridge regression.” Biomedical Imaging (ISBI 2017), 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017).
5. Robinson, Simon Daniel, et al. “An illustrated comparison of processing methods for MR phase imaging and QSM: combining array coil signals and phase unwrapping.” NMR in Biomedicine 30.4 (2017)
6. Dongwook, Lee., et al. "Deep residual learning for compressed sensing MRI.", 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017)
7. Ronneberger, Olaf et al. "U-Net: Convolutional Networks for Biomedical Image Segmentation.", Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015
8. Jenkinson, Mark “Fast, automated, N-dimensional phase-unwrapping algorithm.” Magnetic Resonance in medicine 49.1 (2003): 193-197
9. FSL Prelude: http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FUGUE
10. Visin, Francesco et al., “ReNet: A Recurrent Neural Network Based Alternative to Convolutional Networks”, arXiv:1505.00393
Figures