4690
MRI guided hierarchical sectioning and stitching of brain blocks for alignment of digitized histology to corresponding MR images1Department of Biomedical Engineering, McGill University, Montreal, QC, Canada, 2McConnell Brain Imaging Center, McGill University, Montreal, QC, Canada, 3Montreal Neurological Institute and Hospital, McGill University, Montreal, QC, Canada, 4Department of Neurology and Neurosurgery, McGill University, Montreal, QC, Canada
Synopsis
Ex-vivo MRI of brain tissue can provide important morphological and microstructural information. MR images can also serve as undistorted references for reconstruction of digitized histology, immunohistochemistry or clearing techniques. However, due to space constraints imposed by small animal scanners, imaging a whole human brain generally requires a large bore scanner with limited gradient system performance and constraints on
INTRODUCTION
Ex-vivo imaging of brain tissue using ultrahigh field (UHF) MRI can provide morphological and microstructural information with high spatial resolution. MR images can also serve as undistorted references for registration and reconstruction of tissue volumes processed through histology, immunohistochemistry or clearing techniques. These procedures are important for validation of MRI-based modeling and estimation of microstructural features. However, processing tissue from large brains such as the human brain and comparing to corresponding MRI volumes is not straightforward. One difficulty is that imaging a whole human brain has to be done in a large bore scanner, due to the space constraints imposed by small animal scanners. However, the performance of gradient systems associated with large bore scanners is limited and this limits achievable imaging spatial resolution. Importantly, from a histology standpoint, large tissue sections cut from macaque and human brain are prone to distortions, tears and folding1.
Due to their associated powerful gradient systems and higher main magnetic field, pre-clinical MRI scanners generally provide increased spatial resolution and signal-to-noise ratio (SNR), superior to what can be achieved with large bore human MRI scanners. However, the size of the sample that can be imaged with pre-clinical systems is limited. Here we present a method for MRI-guided hierarchical sectioning and stitching of brain blocks for alignment of processed tissue to corresponding MRI volumes.
The stitching utilizes spatial priors on cut locations, planned using an initial low-resolution MRI. We demonstrate our proposed methodology by sectioning and stitching tissue blocks from a whole hemisphere of a macaque monkey brain.
METHODS
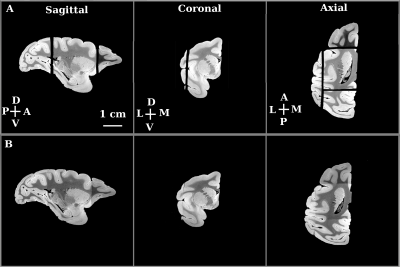
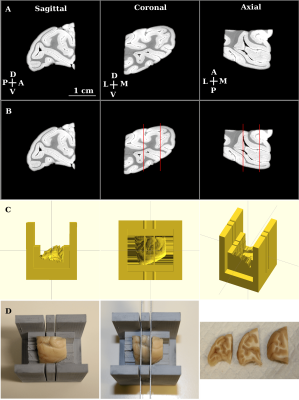
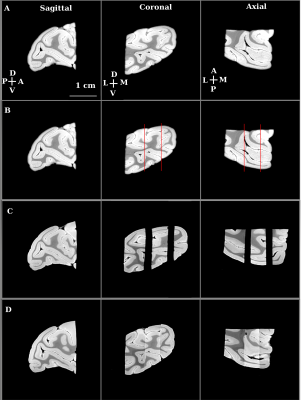
We first imaged a whole, fixed macaque monkey brain ex-vivo using 3 Tesla MRI (Siemens Prisma, SPACE, T2-weighted imaging, voxel-size 600 microns). We then separated the two hemispheres with the aid of a brain specific 3D-printed mold2,3. This separation formed the first stage in the hierarchical sectioning. In the second stage, we used the 3 Tesla MRI data to design a 3D-printed cradle mold that supported the hemisphere during its sectioning into 4 blocks (Figure 1). The 4 sectioned blocks were imaged separately in a small animal scanner (Bruker 7 Tesla PharmaScan; steady-state free precession sequence (SSFP), weighted by the ratio of T2/T1, voxel-size 200 micron). Except for the field of view, the imaging sequence and parameters used for each tissue block were kept unchanged. In the third stage, we used the 7 Tesla data to create a 3D-printed mold for each of the 4 blocks. Figure 4 demonstrates the planning of the mold used for cutting the posterior block (yellow arrows in Figure 1) to 3 slabs. Each slab was imaged separately at 7 Tesla (SSFP, voxel-size 100 micron; Figure 5C). It was then embedded in paraffin, sectioned into slices and stained with Nissl body histology.
The stitching process (Figure 2) is applied to high-resolution MRI volumes of adjacent tissue slabs using spatial priors that identify the surface adjacent to the cut. Adjacent blocks are initially registered using rigid body transformation. Next, a non-linear registration with multi-resolution B-spline is applied. Following the registration step, histogram-based matching of intensities to a reference volume is performed and intensity matched volumes are merged to form one reconstructed volume.
RESULTS
We applied our proposed methodology to an MRI volume of a hemisphere of a macaque brain. Following each stitching stage, we computed the DICE overlap coefficients of the stitched/reconstructed volumes with those of the original volumes (0.9761 for the posterior block). The stitched volumes fit well with the original volumes. In addition, the quality of the stitching was assessed qualitatively by inspecting the continuity of images at the junction of the stitched tissue blocks (Figures 3 and 5).
DISCUSSION
Brain specific, 3D printed model-based tissue sectioning has been proposed for coarse alignment of histology volumes to the corresponding MRI volumes2. The subject-level, aligned brain volumes can be registered to brain prototypes and atlases3. Here we demonstrate how our method can also provide spatial priors of cut locations applicable for an automated stitching algorithm. Moreover, the methodology we propose can be applied hierarchically, enabling stitching of high-resolution small slabs to a whole reconstructed brain. The small slabs can be processed with clearing and staining techniques or with traditional sectioning of slices and staining. The processed tissue slabs can be aligned to the high-resolution MRI slabs, and can be reconstructed to a whole processed tissue brain using the transformations determined by stitching the corresponding MRI slabs.CONCLUSION
Our proposed methodology enables the reconstruction of high-quality digital histology tissue volumes of large brains, including healthy and diseased human brains.Acknowledgements
This research was undertaken thanks to funding from the Canada First Research Excellence Fund awarded to McGill University for the Healthy Brains for Healthy Lives initiative and from the Québec’s Bio-imaging Network.References
1.Amunts, Katrin, Claude Lepage, Louis Borgeat, Hartmut Mohlberg, Timo Dickscheid, Marc-Étienne Rousseau, Sebastian Bludau, et al. 2013. BigBrain: An Ultrahigh-Resolution 3D Human Brain Model. Science 340 (6139): 1472–75.
2.Boopathy Jegathambal, Sethu K, Kelvin Mok, David A. Rudko, Amir Shmuel. MRI based brain-specific 3D-printed model aligned to stereotactic space for registering histology to MRI. 40th IEEE International Engineering in Medicine and Biology Conference, Honolulu, Hawaii, July 17-21, 2018.
3.Luciano, Nicholas J., Pascal Sati, Govind Nair, Joseph R. Guy, Seung-Kwon Ha, Martina Absinta, Wen-Yang Chiang, et al. 2016. Utilizing 3D Printing Technology to Merge MRI with Histology: A Protocol for Brain Sectioning. Journal of Visualized Experiments: JoVE, no. 118 (December). https://doi.org/10.3791/54780.
Figures