4681
Rapid Prototyping of Spiral Based Three-Points Dixon Acquisition and Reconstruction Using PulseqMarina Manso Jimeno1, Sairam Geethanath2, and John Thomas Vaughan2
1University of Groningen, Groningen, Netherlands, 2Columbia University in the city of New York, New York City, NY, United States
Synopsis
Spiral imaging possesses special characteristics that would benefit both research and clinical MR fields. However, its use is often limited due to the difficulties its implementation on a scanner entails. This problem frequently limits the spread and evolution of spirals and consequently their potential and multiple applications. The current study proposes a solution to this problem by demonstrating a rapid prototyping of a three-points Dixon spiral sequence. Results agree with the standard sequence used by the system manufacturer for the same purpose. The software used is open-source and makes possible the sequence implementation in multiple vendor scanners.
Introduction
Spiral trajectory has the benefit of more efficient coverage of k-space compared to the standard Cartesian methods1. Among other advantages, these scans allow for fast acquisitions and improved motion tolerance2. Such characteristics could be favorable in numerous clinical MR-scenarios. However, customized design and implementation of spiral-based whole body sequences on a clinical scanner have typically remained prohibitive. The main reasons are the vendor-specific programming expertise and involved reconstruction techniques required3. The result is a significant increase in invested development time and efforts. Therefore, the goal of this study was to develop and deploy rapid prototyping acquisition and reconstruction methods to overcome this limitation. The demonstration consists of of a three-point Dixon spiral sequence using vendor agnostic and open-source software. Consequently, several MR-applications would benefit not only from the easy accessibility of such sequence but also from the inherent spiral trajectory advantages and Dixon water-fat separation technique.Methods
The acquisition sequence corresponds to a Steady State Free Precession (SSFP), three-point Dixon, golden angle Variable Density Spiral (VDS). Figure 1 provides information about the specific acquisition parameters and the pulse sequence diagram. The study uses Pulseq tool for the sequence design step. This software is MATLAB based, open-source and allows for rapid sequence implementation on multiple vendors through interpreter modules3,4. Automatic python NuFFT-based reconstruction occurs on the scanner. Implementation of standard Dixon methods5 on the resulting complex in-phase and out-of-phase images followed by fat deblurring6 result in water and fat separated images. In vitro experiments utilized two phantoms for sequence testing and validation: the ADNI phantom was used to assesses image quality while water and oil phantoms evaluated tissue separation. Proton Density Fat Fraction (PDFF), a routinely used7 method for fat quantification is additionally estimated as an application validation. In vivo measurements included imaging the thighs of a healthy volunteer. All experiments were performed on a 3T Siemens Prisma scanner and the vendor provided Volume-Interpolated Breath-hold Examination (VIBE) sequence was used for Dixon performance comparison.Results
Figure 2 shows the assessment of the proposed sequence on image quality. Apart from visual inspection, performance tests include SNR and image resolution quantifications. The images provide a SNR of 10 dB and FWHM profile resolution that closely matches the phantom manual. Figures 3 and 4 report the results regarding tissue separation for both the phantom and in vivo experiments. Water and fat separation concur for the proposed and gold standard sequences. Additionally, Figure 5 displays the PDFF analysis comparison for both VIBE and the implemented spiral sequence.Discussion
Results from rapid prototyping experiments showed water and fat separation as expected and in correlation with VIBE. Fat fraction images also provide closely similar results. The main difference lies in the acquisition resolution. Spiral images look blurry possibly because of a phase accumulation in the long read-out direction, caused due to off-resonance. Nevertheless, methods are available to eliminate or palliate these effects. Current and future work involves these corrections to derive accuracy and precision measurements. The challenges of breath-held examination in body imaging studies or whole-body water/fat quantification could be addressed through this implementation. Moreover, an ongoing project on MR-safety uses the proposed sequence as a whole-body pre-scan for temperature map simulations. The current study, provides a rapid prototyping solution to the challenges of designing, implementing and reconstructing non-Cartesian trajectories like spirals. Similarly, implementation is also possible on other Pulseq compatible scanners (GE and Bruker).Conclusion
The study provides evidence on the feasibility of rapidly developing and translating a VDS Dixon sequence to practice. Thus, proving the accessibility of the proposed sequence. The open-source and vendor agnostic characteristics of the software used allow the application of further and customized optimizations, also in multiple manufacturers scanners. These outcomes could facilitate the application of spirals in multiple fields such as MR safety, water-fat tissue measurements and health screening of large populations, among others.Acknowledgements
No acknowledgement found.References
- J. H. Lee, B. A. Hargreaves, B. S. Hu, and D. G. Nishimura. Fast 3D Imaging Using Variable-Density Spiral Trajectories with Applications to Limb Perfusion. Magn. Reson. Med. 2003;50(6):1276–1285.
- B. M. A. Delattre, R. M. Heidemann, L. A. Crowe, et al. Spiral demystified. Magn. Reson. Imaging. 2010;28(6):862-881.
- K. J. Layton, S. Kroboth, F. Jia, et al. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn. Reson. Med. 2017;77(4):1544-1552.
- R. Keerthi Sravan, S. Potdar, P. Poojar, et al. Pulseq-Graphical Programming Interface: Open source visual environment for prototyping pulse sequences and integrated magnetic resonance imaging algorithm development. Magn. Reson. Imaging. 2018;52:9-15.
- G. H. Glover. Multipoint dixon technique for water and fat proton and susceptibility imaging. J. Magn. Reson. Imaging. 1991;1(5):521-530.
- H. Moriguchi, J. S. Lewin, and J. L. Duerk. Dixon Techniques in Spiral Trajectories with Off-Resonance Correction: A New Approach for Fat Signal Suppression Without Spatial-Spectral RF Pulses. Magn. Reson. Med. 2003;50(5):915-924.
- Reeder SB, Hu HH, Sirlin CB. Proton density fat-fraction: a standardized MR-based biomarker of tissue fat concentration. J Magn Reson Imaging. 2012;36(5):1011-4.
Figures

A) Acquisition parameters. B)
Pulse sequence diagram: five 'dummy' pulses lead to SSFP followed by three
spiral acquisitions with different echo times ,
and ,
respectively.

A,B) Top and lateral pictures of the ADNI
phantom. C) Gradient-Echo (GRE) image of the ADNI phantom using the same
acquisition parameters. D) Image of the same slice of the ADNI phantom acquired
with the proposed sequence.

A) Pictures of the water and oil phantoms. B) Water and
fat-only images acquired with VIBE and the same acquisition parameters. C)
Water and fat-only images acquired with the proposed sequence. Note that the
water image is subjected to chemical shift and gradient delay artifacts.
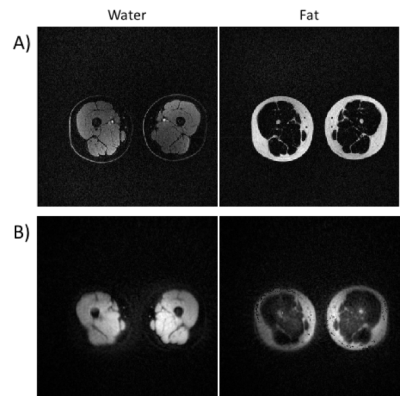
In vivo imaging of the thighs. A) Water and
fat-only images acquired with VIBE and the same acquisition parameters. B)
Water and fat-only images acquired with the proposed sequence. Note that the
outermost skin layer is not perceivable in the water image.
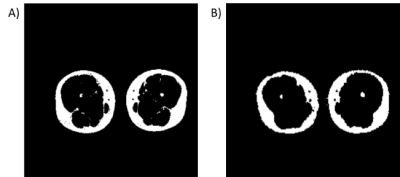
Fat fraction images (Fat/(Water+Fat)) for A) VIBE and B)
the proposed sequence.