4398
DeepBLESS: learning inverse Bloch equations for rapid prediction of myocardial relaxation parameters1Department of Radiological Sciences, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, United States, 2Department of Medicine, Division of Cardiology, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, United States, 3Division of Cardiology, Veterans Affairs Greater Los Angeles Healthcare System, Los Angeles, CA, United States, 4Biomedical Physics Inter-Departmental Graduate Program, University of California, Los Angeles, Los Angeles, CA, United States
Synopsis
Bloch equation simulation provides accurate estimation of soft tissue relaxation parameters for many applications. To speed up using Bloch equation for relaxation parameter estimation, we propose a general approach - deep learning with Bloch equation simulations (DeepBLESS) - to learn inverse Bloch equation for rapid myocardial relaxation parameter prediction. Using the Modified Look-Locker inversion recovery (MOLLI) sequence and a self-designed simultaneous radial T1 and T2 mapping sequence as examples, we demonstrated that DeepBLESS was adaptive to heart rate variation with good estimation accuracy and precision while reducing the inline computation time compared to the conventional Bloch-equation-based approaches.
Introduction
Bloch equation simulation provides accurate estimation of soft tissue relaxation parameters for many applications1-4. However, it is time inefficient when substantial detail is included in the simulations. Deep learning has been used in MR fingerprinting to accelerate parameter estimation by using the acquired signal as input1. For cardiac applications however, incorporating the acquired signal is more challenging because the signal acquisition time varies with beat-to-beat variation in the heart rate. To solve this issue, we propose a general approach of using deep learning with Bloch equation simulations (DeepBLESS) for rapid myocardial relaxation parameter prediction. Using the Modified Look-Locker inversion recovery (MOLLI) sequence2-4 and a self-designed simultaneous radial T1 and T2 mapping sequence as examples, we will demonstrate that DeepBLESS can be adaptive to heart rate variation with good estimation accuracy and precision while significantly reducing the estimation time compared to conventional approaches using Bloch equation simulations.Methods
Fig.1 illustrates the proposed deep learning model for DeepBLESS. The self-designed radial T1/T2 mapping sequence is explained in Fig.2. Similar to raMOLLI5, compressed sensing with view sharing were used to reconstructed 10 images for each heartbeat, resulting in 110 image signal in total. In DeepBLESS model, the input signal was formatted as an 8×2 matrix (one image signal row and one acquisition time row) for the MOLLI 5-(3)-3 sequence and was formatted as an 11×11 matrix (ten acquired signal + one time signal as a column for each heartbeat) for the simultaneous radial T1-T2 mapping sequence.
Bloch equation simulations were used to generate the training sets (1,000,000 samples) and validation sets (100,000 samples) for each sequence (the MOLLI or radial T1/T2 mapping sequence). Random T1, flip angle α and heart rate (HR) were uniformly sampled for the range 200 ms – 2000 ms, 20°- 45° for MOLLI / 3° - 8° for the radial sequence, and 40 bpm -100 bpm, respectively. For T2, 90% was uniformly random-sampled between 20 ms – 100 ms and 10% was uniformly random-sampled between 100 ms – 200 ms. For each group of T1, T2, α, and HR, 10% Gaussian noise was added to each heartbeat interval before simulation. For the MOLLI/radial sequence, either 1% or 5% Gaussian noise was respectively added to the simulated signal. For training, we used mean square error (MSE) as the loss function with a batch size of 2000. Model parameters with the best MSE from the validation set were saved and used for phantom and in vivo studies.
After training, DeepBLESS was evaluated and compared with a conventional Bloch equation-based-approach (BLESSPC3) using phantoms at simulated heart rates of 40 - 100 bpm and in 8 healthy volunteers for MOLLI at 1.5T and in 10 healthy volunteers for the simultaneous radial T1-T2 mapping sequence at 3.0 T. Two-tailed Student’s t-tests were used for pair-wise comparisons. Both the inversion and T2 preparation pulses were simulated in detail for the radial T1-T2 mapping sequence, while for MOLLI, a fixed inversion factor of 0.963 was used.
Results
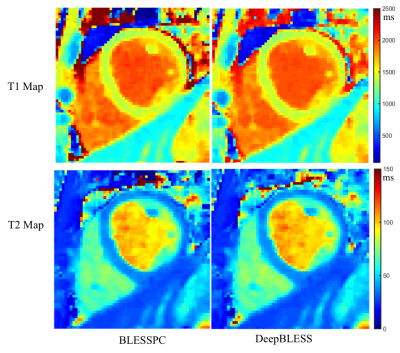
In phantom studies, DeepBLESS and BLESSPC achieved similar accuracy and precision in T1/T2 estimations for both the MOLLI and radial T1-T2 mapping sequences (p > 0.05), as shown in Table 1. At in vivo, DeepBLESS and BLESSPC generated similar myocardial T1/T2 values (p > 0.05) and precision and repeatability (p> 0.05), as shown in Table 2. The time to generate an in vivo T1 or T2 map using DeepBLESS was < 1s, while using BLESSPC takes ~98 sec for MOLLI and ~ 3 hours for the radial T1-T2 mapping sequence. Figure 3 and 4 show examples of myocardial T1/T2 maps generated using DeepBLESS and BLESSPC for the MOLLI and radial T1-T2 mapping sequences, respectively.Discussion
The proposed DeepBLESS approach enabled in-line estimation of myocardial relaxation parameters to be almost instantaneous and removed the rate-limiting step for estimation of relaxation parameters by off-loading the time-consuming task of Bloch equation simulations to off-line. Phantom and in vivo results showed that DeepBLESS can achieve 490 – 18,000 times acceleration compared to BLESSPC with similar accuracy and precision.
Conclusion
DeepBLESS offers an almost instantaneous approach for estimating relaxation parameter maps with good accuracy and precision compared to the conventional Bloch equations-based approaches. The acceleration provided by DeepBLESS is promising for multi-parametric mapping.Acknowledgements
NoneReferences
1.Cohen et al. MRM, 2018;80(3):885-94.
2. Messroghli et al. MRM, 2004; 52(1):141-146.
3. Shao et al. MRM, 2017;78(5):1746-1756.
4. Christos et al. JCMR, 2015;17:104.
5. Marth et al. MRM, 2018;79:1387-1398.
Figures