3836
MRI Driven Augmented Reality Image-Guidance Platform for Myocardial Cell Delivery1Medical Biophysics, University of Toronto, Toronto, ON, Canada, 2Schulich Heart Research Program, Sunnybrook Research Institute, Toronto, ON, Canada, 3McEwen Centre for Regenerative Medicine, University Health Network, Toronto, ON, Canada
Synopsis
Myocardial infarction is a leading cause of heart failure, a condition with a 5-year mortality rate of ~50%. Current HF management techniques aim to slow disease progression rather than improve function. Cardiac regenerative medicine through cell-based approaches offers the potential to repopulate non-contractile scar tissue. However, regions of scar are not well delineated during the cell-delivery procedures, and accurate cell placement is vital to realize positive outcomes from therapy. We aim to develop a novel integrative image-guidance solution combining MRI for myocardial scar characterization with augmented reality via a head-mounted display to visualize critical MRI information intraoperatively and guide delivery.
Introduction
Acute myocardial infarction (MI) remains the leading cause of heart failure (HF), with 25% of survivors developing HF, a condition with a 5-year mortality rate of ~50% 1. Current HF management techniques focus on slowing disease progression rather than improving contractile function. Cardiac regenerative medicine (CRM) using cell-based approaches, offers a potentially revolutionary mechanism to repopulate non-contractile scar tissue and overcome the limited regenerative capacity of the human heart 2,3. To realize improvements in cardiac function it is vital that cells are delivered precisely to regions of scar tissue, minimizing incorrect targeting to surrounding healthy tissue, myocardial perforation and systemic embolization 5. However, during an open-chest surgical cell injection procedure, the infarcted territory is not is not well delineated for accurate targeting. Due to its superior soft tissue contrast, MRI is considered the gold standard for cardiac function and scar viability imaging post MI 4. We aim to develop a novel integrative solution to guide cell injections using late gadolinium enhancement (LGE) cardiac MRI to generate 3D scar roadmaps and augmented reality (AR) guidance through an optical see-through head-mounted display (OST-HMD) to visualize critical MR imaging information intraoperatively.Methods
Device: We have utilized the Microsoft HoloLens OST-HMD for initial experiments due to its class leading performance in contrast perception, task load and frame rate 5. Our initial implementation leverages the HoloLens front facing sensor capabilities for pose estimation through robust square-based fiducial tracking 6. First, a standard camera calibration procedure was performed through recording multiple views of a known planar chessboard pattern to estimate camera intrinsic parameters. Second, to compensate for positional errors common to OST-HMDs, and arising from the differing coordinate systems of the user’s eyes and the front facing sensors 5, a transform from the sensor stream frame to the user’s vision frame of reference is required. A modified version of the single-point active alignment method was implemented to compute this transform and minimize discrepancy in virtual model registration for individual users 7.
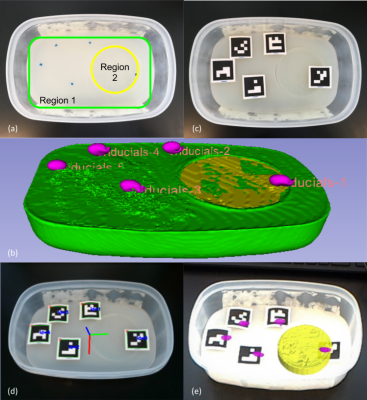
Phantom study: An agar phantom was prepared to provide a substrate for future mock injection studies. The phantom is comprised of two regions with differing T1 contrast, representing the organ and an embedded target. Vitamin E tablets within the agar gel served as MRI-visible fiducial markers, which are later aligned with custom square-based fiducial landmarks to enable registration between AR projections of the MRI volumes and the real scene. The phantom was imaged using a 3D-FSPGR sequence on a 3T whole-body scanner.
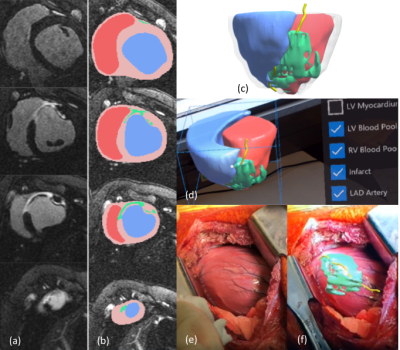
Animal study: Using a porcine MI model (N=3), infarcts were characterized through late gadolinium enhanced (LGE) MRI and infarcts were segmented using a full width half maximum algorithm to semi-automatically quantify infarcted areas 8. Models were optimized for viewing through color-coding and transparency schemes to identify individual structures, and saliency techniques to highlight infarct regions.
Results
Phantom study: The agar phantom is shown in Figure 1a, alongside a segmented 3D model from the FSPGR dataset, created using 3D Slicer 9 (Figure 1b). Square fiducial markers are aligned with the MRI fiducial locations on the phantom (Figure 1c), creating consistent landmarks between preoperative and intraoperative image space. Robust visual tracking of the custom marker configuration using the front-facing sensors on the HoloLens permitted virtual model alignment with the intraoperative scene with reasonable accuracy (Figure 1d, e).
Animal study: 3D MRI models of the heart and scar anatomy (Figures 2a-c) were displayed to the user through the HoloLens device alongside interfaces for interaction (Figure 2d). The surgeon’s point of view for an open chest cell-injection procedure is shown in Figure 2e, f, with a virtual 3D scar roadmap representing the extent of myocardial scar tissue included.
Conclusion
Our study offers the first proof-of-concept MRI-guidance solution for accurate surgical delivery of cell-based therapies to the heart. Initial results of OST-HMD guidance are promising; however, more work is necessary to ensure consistent virtual content registration stability and accuracy. Our system could improve on a challenging aspect of cell delivery, potentially leading to improved management of HF. Future work is focused on the development of improved per-user device calibration methods and motion tracking of moving heart structures.Acknowledgements
Ontario Research Fund (ORF-RE7-21), Ontario Institute for Regenerative Medicine, Medical Biophysics - University of Toronto.References
1. Heart and Stroke. The burden of heart failure. Heart and Stroke (2016). Available at: https://www.heartandstroke.ca/-/media/pdf-files/canada/2017-heart-month/heartandstroke-reportonhealth-2016.ashx?la=en. (Accessed: 9th March 2018)
2. Gerbin, K. A. & Murry, C. E. The winding road to regenerating the human heart. Cardiovascular Pathology 24, 133–140 (2015).
3. Chong, J. J. H. et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 510, 273–277 (2014).
4. Pop, M. et al. Quantification of fibrosis in infarcted swine hearts by ex vivo late gadolinium-enhancement and diffusion-weighted MRI methods. Physics in Medicine and Biology 58, 5009–5028 (2013).
5. Qian, L. et al. Comparison of optical see-through head-mounted displays for surgical interventions with object-anchored 2D-display. International Journal of Computer Assisted Radiology and Surgery 12, 901–910 (2017).
6. Garrido-Jurado, S., Muñoz-Salinas, R., Madrid-Cuevas, F. J. & Marín-Jiménez, M. J. Automatic generation and detection of highly reliable fiducial markers under occlusion. Pattern Recognition 47, 2280–2292 (2014).
7. Azimi, E., Qian, L., Navab, N. & Kazanzides, P. Alignment of the Virtual Scene to the 3D Display Space of a Mixed Reality Head-Mounted Display. arXiv:1703.05834 [cs] (2017).
8. Pop, M. et al. High-Resolution 3-D T$\bf _1^\bf *$-Mapping and Quantitative Image Analysis ofGRAY ZONEin Chronic Fibrosis. IEEE Transactions on Biomedical Engineering 61, 2930–2938 (2014).
9. Kikinis, R., Pieper, S. D. & Vosburgh, K. G. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. in Intraoperative Imaging and Image-Guided Therapy (ed. Jolesz, F. A.) 277–289 (Springer New York, 2014). doi:10.1007/978-1-4614-7657-3_19
Figures