3397
Motion Compensation for Free-Breathing Diffusion-Weighted Imaging (MoCo DWI)1Division of Medical Physics in Radiology, German Cancer Research Center (DKFZ), Heidelberg, Germany, 2Faculty of Physics and Astronomy, Heidelberg University, Heidelberg, Germany, 3Siemens Healthcare GmbH, Erlangen, Germany, 4Medizinische Fakultät Heidelberg, Heidelberg University, Heidelberg, Germany
Synopsis
Diffusion-weighted imaging (DWI) of the abdomen has acquisition times of several minutes. For this reason respiratory motion can cause misalignment between acquired slices at the same position but different b-values. To overcome this, we estimate the patients’ respiratory motion using a T1-weighted, stack-of-stars GRE pulse sequence and an advanced 4D reconstruction. This motion estimation is used to compensate for respiratory motion in a common, free-breathing DWI acquisition. In three volunteers an improved alignment of structures in the liver are shown. This allows for a better comparison and potential benefits for further processing (e.g. for ADC-maps).
Introduction
Diffusion-weighted imaging (DWI) of the abdomen is a valuable diagnostic tool, e.g. for detection and characterization of liver lesions and treatment response assessment. Due to acquisition durations of several minutes, respiratory motion can cause misalignment of slices acquired at different points in time. To prevent this, breath-hold or prospective triggering can be used during data acquisition1. Alternatively, it is possible to avoid these artifacts from free-breathing scans by retrospective gating2,3 or motion compensation4. Breath-hold acquisitions are not feasible for all patient groups, and prospective triggering prolongs the acquisition duration roughly two-fold. Retrospective gating suffers from low signal-to-noise-ratio (SNR) efficiency and can lead to missing slices in 3D volumes which need further processing2.
Current motion compensation methods in the abdomen are limited to affine motion4 which is not the case for liver motion induced by respiration. Furthermore, image-based deformable motion detection in DWI is challenging as the images primarily show areas of restricted diffusion and SNR decreases with increasing b-value4. This challenge is even increased for the typical axial slice orientation with a slice-thickness of 5 to 7 mm as this means low resolution in the main motion direction. Therefore, we propose a novel approach which uses an independent stack-of-stars gradient-echo (GRE) sequence – that is free from these obstacles – to estimate the respiratory motion. This estimation is then used to compensate the motion during image reconstruction of a free-breathing DWI acquisition.
Methods
Three volunteers were scanned on a 1.5 T MRI scanner (MAGNETOM Aera, Siemens Healthcare, Erlangen, Germany). The golden-angle stack-of-stars prototype pulse sequence5 was used with the joint-MoCo-HDTV reconstruction algorithm6 for the motion estimation (radial views = 1300, TA = 287 s, FA = 12°, TR = 3.7 ms, FOV = 385 × 385 × 395 mm³, matrix = 256 × 256 × 80). Sagittal slice orientation ensures high resolution in the main motion direction for optimal motion estimation7. Additionally, 35 axial DWI slices were acquired with a prototype single-shot echo-planar imaging sequence for b = 50, 400, 800 s/mm² with 8, 8, and 16 averages, respectively (TA = 227 s, FOV = 378 × 307 × 204 mm³, matrix = 256 × 208 × 35).
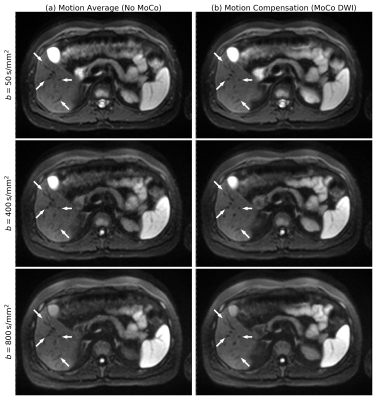
Three different post-processing algorithms of the DWI scan are compared. (a) No motion compensation (No-MoCo); the measurements are averaged for each b-value without any consideration of motion. (b) Retrospective gating assigns independently one of 10 motion phases to each DWI slice based on a respiratory cushion. Slices at the same position and motion phase are combined by averaging. Only the end-exhale phase is kept. (c) Motion-compensated DWI (MoCo DWI) uses the retrospectively gated images as just described (but all motion phases) and deforms them to the end-exhale phase using the previously estimated motion. These 10 volumes are then combined to a single volume. The whole workflow is sketched in Figure 1.
We evaluate the position of vessels within the liver to assess the effect of the motion compensation. The vessels function as replacement for lesions which do not exist in the healthy volunteers we measured. In each volunteer two different vessels in different slices are manually segmented in all three b-values. The pairwise dice coefficients are used to compare the alignment.
Results
The No-MoCo images show displacements of vessels between different b-values. The retrospective gating suffers from low SNR especially in the higher b-values, signal voids due to phase cancelations and holes in several phases (Figure 2). These disadvantages are solved in the MoCo DWI which has higher SNR and no observable cancelations in any of the volunteers (Figure 3). The superior-inferior motion of the liver was (6.9 ± 0.3) mm at its highest point and thus only slightly larger than the DWI slice-thickness. The dice coefficients showed a relative increase of 8 %, 17 % and 43 % for the three volunteers.Discussion and Conclusion
In this study we successfully transferred motion estimations from a GRE to a DWI pulse sequence. The drawback of MoCo DWI is the requirement of the additional GRE sequence which prolongs the measurement. However, this is required because the motion in superior-inferior direction is similar to the slice-thickness of the diffusion-weighted images. For this reason proper motion estimation on diffusion-weighted images is not feasible. This proof-of-concept study of MoCo DWI shows improved alignment of structures between b-values. This promises improved delineation and detection particularly of small lesions. In further work, we will evaluate the impact of the improved spatial alignment on ADC maps and extend the study to patient cases.Acknowledgements
Parts of the reconstruction software were provided by RayConStruct GmbH, Nürnberg, Germany.References
- H. Chandarana and B. Taouli: Diffusion and perfusion imaging of the liver. European Journal of Radiology 76(3), 348 (2010).
- T. N. van de Lindt, M. F. Fast, U. A. van der Heide, and J.-J. Sonke: Retrospective self-sorted 4D-MRI for the liver. Radiotherapy and Oncology 127(3), 474 (2018).
- Yilin Liu, Xiaodong Zhong, Brian G. Czito, Manisha Palta, Mustafa R. Bashir, Dale Brian M., Yin Fang‐Fang, and Cai Jing: Four‐dimensional diffusion‐weighted MR imaging (4D‐DWI): a feasibility study. Medical Physics 44(2), 397 (2017).
- Y. Mazaheri, R. K. G. Do, A. Shukla-Dave, J. O. Deasy, Y. Lu, and O. Akin: Motion Correction of Multi-b-value Diffusion-weighted Imaging in the Liver. Academic Radiology 19(12), 1573 (2012).
- K. T. Block, H. Chandarana, S. Milla, M. Bruno, T. Mulholland, G. Fatterpekar, M. Hagiwara, R. Grimm, C. Geppert, B. Kiefer, and D. K. Sodickson: Towards Routine Clinical Use of Radial Stack-of-Stars 3D Gradient-Echo Sequences for Reducing Motion Sensitivity. Journal of the Korean Society of Magnetic Resonance in Medicine 18(2), 87 (2014).
- C. M. Rank, T. Heußer, M. T. A. Buzan, A. Wetscherek, M. T. Freitag, J. Dinkel, and M. Kachelrieß: 4D respiratory motion-compensated image reconstruction of free-breathing radial MR data with very high undersampling. Magn. Reson. Med. (2016).
- R. Grimm, S. Fürst, M. Souvatzoglou, C. Forman, J. Hutter, I. Dregely, S. I. Ziegler, B. Kiefer, J. Hornegger, K. T. Block, and S. G. Nekolla: Self-gated MRI motion modeling for respiratory motion compensation in integrated PET/MRI. Medical Image Analysis 19(1), 110 (2015).
Figures