3353
Estimation and Correction of Image Phase in diffusion weighted MRI using Deep Learning1Computer Science, Technical University Munich, Munich, Germany, 2GE Healthcare, Munich, Germany, 3Physics, Technical University Munich, Munich, Germany, 4MR Applied Science Laboratory Europe, GE Healthcare, Stockholm, Sweden
Synopsis
The phase of diffusion weighted MR images (DWI) is regularly discarded in clinical application although it might contain valuable information, as it is composed of phase contributions due to rigid motion, eddy currents and brain pulsation among others.
In this work we take advantage of a neural network to separate the different phase components in individual DWI phase images. This enables estimating the amount of brain pulsation from DWI and modelling brain pulsation. The gained information may be used for phase correction, which eventually will allow using real-valued DWI (instead of magnitude DWI) to eliminate the Rician bias.
Introduction
The clinical application of diffusion weighted MR imaging mostly relies on the magnitude of the complex MR signal only. However, removing the phase has gained recent attention1-3 as the phase of diffusion weighted MR images (DWI) is additively composed of several phase components and contains relevant information3: $$\phi = \phi_0 + \phi_{\chi}+\phi_{eddy} +\phi_{motion,rigid} +\phi_{motion,nonrigid}$$
Recently we proposed an analytical pipeline for Diffusion Spectrum Imaging (DSI) acquisitions taking advantage of q-space symmetry, which separates phase components resulting from eddy currents, bulk motion and allows determining the phase component, which results from intracranial motion only. Sprenger’s approach however requires an extensive DWI dataset to determine the non-linear eddy currents, and thus leaves room for improvement regarding the computational cost and robustness.
Here we separate the phase component due to eddy currents and brain pulsation in DWI taking advantage of a deep learning approach. We bypass computationally expensive modelling and aim at a robust model applicable to individual DWI phase images.
Methods
We propose a modified UNet architecture4 to estimate the phase component due to brain pulsation from the DWI phase. The network was trained using Adam optimization5, L2 loss, batch normalization, learning rate of 10-4 and dropout rate of 0.3.
Data from seven healthy volunteers were acquired with a 3T GE MR750 scanner (GE Healthcare, Milwaukee, WI) using a 32-channel head coil and Stejskal-Tanner diffusion preparation with maximum b-value of 1000 s/mm2, different diffusion-encoding directions and strengths and single-shot 2D EPI readout, after obtaining written informed consent. Further acquisition parameters were: isotropic resolution of 2.5mm, TR/TE = 1800ms/80.7ms, ESP = 0.592 ms, parallel imaging factor 2 and SENSE reconstruction. The total number of DWI acquisitions in the dataset is 4582.
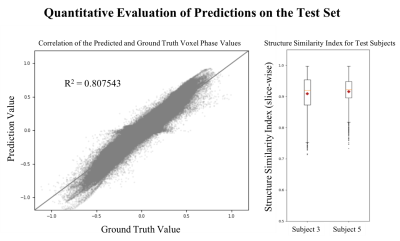
Figure 1 shows the data processing for training and prediction. The phase of the reconstructed DWI signal is preprocessed using phase unwrapping6. The network receives the unwrapped phase images and is trained to predict the phase resulting from brain pulsation. To determine the phase due to brain pulsation, i.e. the ground-truth for the deep learning approach, the analytical pipeline introduced in3 is applied. Prediction quality of the model is assessed using the Structure Similarity Index (SSIM) and the R2-score.
Results
From figure 2 it can be observed how the predictions approximate the ground-truth with increasing quality for both the training and validation set. This is confirmed quantitatively by the decreasing training and validation losses (figure 3).
Figure 4 shows the prediction and ground-truth phase images for representative test samples. Visual inspection of the ground-truth and prediction show a very good agreement. Figure 5 quantitatively shows the agreement of the prediction with the ground-truth for all test samples as well as the distribution of the SSIM.
Discussion
It is observed visually that predicted phase images show very good agreement with the assumed ground-truth. The quantitative measures confirm the excellent prediction capability.
Predictions that show very low (R2 < -0.7) appear to have a systematic error, which we attribute to residual phase errors (incomplete eddy current phase removal) in the ground-truth produced by the analytical approach. In figure 4g it is observed that the ground-truth still includes a linear phase offset which indicates that it shows not only the phase due to brain pulsation.
In future work we will improve prediction quality by increasing the size of the training data. Further we will employ data augmentation to a larger extent, continuing to model eddy current phase contributions as in7 (preliminary data not shown) to improve the network performance.
Conclusion
In this work we successfully demonstrated a method that separates the phase components in DWI due to eddy currents and brain pulsation with excellent agreement (R2 = 0.8). The progress made compared to our previous analytical estimation of the intracranial motion induced phase is the computational efficiency. We emphasize that the novel DL based method works on single two-dimensional DWI instead of a full DSI acquisition.
We see two applications of the proposed method. Firstly, it may contribute to a method that models all phase components such that signal and noise may be distinguished in the phase. Thus, full phase correction may be performed to yield real-valued data, preventing Rician bias. Secondly, the intracranial motion can be determined from flow-induced phase component through the inherent velocity encoding in the DWI acquisition. This may eventually help in understanding the glymphatic system, the diagnosis of tumor induced chances affecting the CSF8 and the diagnosis of hydrocephalus9.
Acknowledgements
No acknowledgement found.References
1. Pizzolato M, Fick R, Boutelier T, Deriche R. Noise Floor Removal via Phase Correction of Complex Diffusion-Weighted Images: Influence on DTI and q-space Metrics. Computational Diffusion MRI. 2016.
2. Pizzolato M, Deriche R. Automatic and Spatially Varying Phase Correction for Diffusion Weighted Images. Proc Intl Soc Mag Reson Med. 2018
3. Sprenger T, Sperl J, Fernandez B, Haase A, Menzel M. Real valued diffusion-weighted imaging using decorrelated phase filtering. Magnetic Resonance in Medicine. 2016.
4. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. arXiv. 2015.
5. Kingma D P, Ba J L. Adam: A method for stochastic optimization. ICLR. 2015.
6. Abdul-Rahman H S, Gdeisat M A, Burton D R, Lalor M J, Lilley F, Moore C J. Fast and robust three-dimensional best path phase unwrapping algorithm. Applied Optics. 2007.
7. Xu D, Maier J, King K, Huang H, Zhao X, Collick B, Wu G, Peters R, Hinks S. Revisiting GRAFIMAGE: Measurement and Characterization of High Order Eddy Currents using a Phantom. GE Healthcare. 2012.
8. De Souza Bezerra M L, Andorinho de Freitas Ferreira A C, de Oliveira-Souza R. Pseudotumor Cerebri and Glymphatic Dysfunction. Front Neurol. 2017.
9. Zhu D, Xenos M, Linninger A, Penn R. Dynamics of lateral ventricle and cerebrospinal fluid in normal and hydrocephalic brains. Journal on Magnetic Resonance Imaging. 2006.
Figures

Figure 1: Visualization of the data flow for training and prediction.
Top: For training, the reconstructed phase image is filtered and unwrapped (a) giving the input image. The phase contributions from linear motion and eddy currents are determined as in [1] and subtracted (b) to create the output image. Input and output are standardized using the mean and standard deviation of the input (c) for training (d).
Bottom: For prediction, the input phase image is pre-processed (e), standardized (f) and passed through the network (g). The prediction resulting from (g) is rescaled and translated using the standardization derived from the input image (h).

Figure 2: Training progress of a random selection of slices and diffusion encoding q-vectors from the training set (a) and validation set (b).
The left column depicts the input phase image. The following columns show the predictions of the network during training. The very right column shows the ground-truth phase image. Both input and output phase are shown in rad.
The approximation improves with the number of iterations. After epoch 25 the network predicts the rough structure of the ground-truth. After epoch 100 fine structures are also recognizable in the prediction in agreement with the ground-truth.