2823
Automated Prediction of Stroke Lesion Outcome using Multiparametric Deep Neural Network1School of Data and Computer Science, Sun-Yat Sen University, Guangzhou, China, 2Academy for Advanced Interdisciplinary Studies, Peking University, Beijing, China, 3YITU Healthcare, Shanghai, China, 4Department of Radiology, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, China
Synopsis
Recent stroke trials raised a demand for triage decision intelligence of ischemic lesion progression. This study aimed to develop a multiparametric deep neural network to segment regions that predicted final infarct formation. The PWI-derived CBF, CBV, MTT and Tmax maps served as multi-channel inputs to algorithm training. We used a 2.5D U-Net to generate lesion segmentation. Our approach showed a good sensitivity and specificity with AUC of 0.868 in predicting the final lesions, and a comparable performance of DICE and IOU. In conclusion, we demonstrated feasibility for predicting tissue outcome in acute ischemic stroke with multiparametric deep learning algorithm
Introduction
The evolution of acute ischemic stroke (AIS) is a highly heterogeneous process. Recent advances in AIS clinical trials (DEFUSE-3, DAWN & EXTEND-IA)1-3 designed a pre-treatment triage strategy using multiparametric imaging-based decision-making tool. Diffusion-weighted (DWI) and perfusion-weighted imaging (PWI) or CT perfusion (CTP) were applied in these trials to identify tissue-at-risk (salvageable penumbra) and quantify temporal profiles of penumbra, infarct and their mismatch. However, the “infarct” detected by DWI may still be reversible. The CTP or PWI-derived time to maximum residue function (Tmax)>6s is used to define tissue-at-risk4, and DWI-derived apparent diffusion coefficient (ADC)<600x10-6 mm2/s is considered as infarct5. The thresholding method may not reflect an objective insight of ischemia progression because different population may have varying tissue physiology that affects collaterals, reperfusion and tissue outcome. Therefore, a demand for decision intelligence of stroke tissue progression is raising. The bloom of machine and deep learning applied in computer vision and pattern recognition has begun to revolutionize the medical diagnosis and image analysis. In this study, we sought to develop a multiparametric deep neural network to segment regions which were predictive of final infarct formation in AIS patients. The multiparametric maps derived from PWI were simultaneously incorporated as inputs of the network to predict follow-up DWI lesion (ground truth).Methods
The patients enrolled in this study were from publicly available cohort database, ischemic stroke lesion segmentation (ISLES) challenge 2018 (http://www.isles-challenge.org). All patients underwent imaging examinations within 8 hours of stroke onset, and DWIs were performed within 3 hours after perfusion imaging. The PWI/CTP-derived cerebral blood flow (CBF), cerebral blood volume (CBV), mean transit time (MTT) and Tmax maps served as multi-channel inputs to algorithm training. The ischemic regions with hyperintensity on the DWIs were manually drawn as infarction by experienced radiologists, and used as annotated ground truth. The data were divided to two sets (training = 86 cases and validation = 8 cases).
The algorithm was implemented using PyTorch library and executed on GeForce GTX 1080Ti GPU. For this specific task, we designed a two-step algorithm module, lesion identification layer and segmentation layer. For lesion identification, the annotated multiparametric maps were trained to detect lesions on all image slices. The resulting outputs of positive and negative layers were subsequently trained by an U-Net to generate pixel-wise confidence score for the infarct segmentation (Figure 1). We introduced a 2.5D U-Net as segmentation backbone, combining the consecutive slices superior to and inferior to the lesions of interest as overall inputs. The data augmentations (random vertical/horizontal flips and rotations) were applied to avoid overfitting. The learning was optimized by Adam stochastic optimization with back-propagation method at the learning rate of 0.001, iteratively minimizing the loss function between the model output and ground truth. The cross entropy and DICE score were used as loss function for the identification and segmentation layer, respectively. The performance of the algorithm was evaluated on the validation set with average DICE, intersection over union (IOU), and area under the receiver operating characteristic curve (AUC), case-level sensitivity and specificity
Results
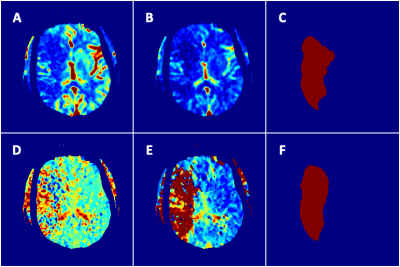
The performance of infarct prediction indicated a mean IOU of 0.397±0.150 and mean Dice score of 0.552 ±0.150. The discriminating power of infarct from non-infarct area was measured at case-level, including a mean sensitivity of 0.743 ±0.074, mean specificity of 0.993 ±0.004, and mean AUC of 0.868 ±0.037. An illustration of tissue outcome prediction was shown using the multiparametric deep neural network (Figure 2).Discussion
The algorithm showed a moderate to good performance in predicting tissue outcome. The similarity (DICE) and overlap (IOU) of the predicted outcome to the ground truth showed a moderate performance (comparable to the 1st place of ISLES challenge 2018), whereas case-level AUC, sensitivity and specificity for final lesion prediction demonstrated a much better performance. The plausible reason could be multifactorial such as collateral flow and size of reperfusion area that play important roles in lesion growth. In addition, therapeutic effect was not considered in the modeling since no such information was available. The visual features extracted from 2.5D U-Net need to be carefully selected for training. We used multiparametric perfusion maps as the training inputs, but these post-processed maps may not represent authentic and underlying features of ischemic pathophysiology due to variation from different processing algorithms and noise. The future improvement should focus on training of 4D source images from PWI or CTP, because more dynamic rather than static information can be retrieved from 4D signal domainConclusion
Our study revealed the feasibility for predicting tissue outcome in AIS with multiparametric deep learning algorithm, and further investigation will focus on decoding potential correlation with individualized clinical outcome.Acknowledgements
No acknowledgement found.References
1. CAMPBELL, B.CV, et al. Endovascular therapy for ischemic stroke with perfusion-imaging selection. New England Journal of Medicine, 2015, 372.11: 1009-1018.
2. NOGUEIRA, R G., et al. Thrombectomy 6 to 24 hours after stroke with a mismatch between deficit and infarct. New England Journal of Medicine, 2018, 378.1: 11-21.
3. ALBERS, G W., et al. Thrombectomy for stroke at 6 to 16 hours with selection by perfusion imaging. New England Journal of Medicine, 2018, 378.8: 708-718.
4. CAMPBELL, B.CV, et al. Comparison of computed tomography perfusion and magnetic resonance imaging perfusion-diffusion mismatch in ischemic stroke. Stroke, 2012, 43.10: 2648-2653.
5. STRAKA, M, et al. Real‐time diffusion‐perfusion mismatch analysis in acute stroke. Journal of Magnetic Resonance Imaging, 2010, 32.5: 1024-1037.