2515
Measurement of 23Na MRI point-spread function (PSF) using a 3D printing resolution phantom1School of Biomedical Engineering, McMaster University, Hamilton, ON, Canada, 2Imaging Research Centre, St. Joseph's Healthcare, Hamilton, ON, Canada, 3GE Global Research, Munich, Germany
Synopsis
Acquisition of in vivo 23Na MRI suffers from many inherent technical challenges, including low signal, short T2 relaxation times, and the necessity of dedicated hardware for transmitting and receiving. Despite these issues, research remains attractive because of sodium’s essential role in cellular homeostasis, pH regulation and action potentials in neurons. Quantification of data acquisition and reconstruction techniques are essential in order to overcome 23Na MRI’s difficulties, and we present measurement of the point-spread functions of 3D radial pulse sequences in resolution phantoms with differing sodium concentrations.
Purpose
In vivo 23Na MRI is desirable due to sodium's essential role human metabolism,1,2 but its acquisition suffers from many inherent technical challenges. Among these difficulties are the low gyromagnetic ratio, the requirement of dedicated transmit/receive coils, non-standard pulse sequences, low signal and henceforth long acquisitions.3,4 Ultrashort echo time (UTE) pulse sequences are preferred in order to capture the signal from the short T2 times in 23Na MRI, and among the most common of these is 3D radial projection (3DRP) imaging.5 In vivo brain sodium concentrations can range from 15 mM (bound intracellular sodium) to 150 mM (cerebrospinal fluid), thereby necessitating the imaging and reconstruction methods to be sensitive to a broad signal range. We present here quantitative measurement of modulation transfer functions and point-spread functions (PSFs) in a saline resolution phantom for different concentrations of sodium with 3DRP pulse sequences.
Methods
The
imaging experiments were conducted using a GE 3T MR750 (General
Electric Healthcare, Milwaukee, WI), using a custom designed/built
single-tune birdcage head coil (resonant frequency = 33.7 MHz). A phantom (see Figure 1) was 3D-printed and filled with different concentrations of NaCl and distilled water (150, 60, and 30 mM), before imaging in the coronal plane. A single healthy volunteer was also scanned in order to qualitatively assess the performance for neuroimaging.
Imaging was conducted using the GE MNS Research Pack (v. 2018-07-18). 3DRP UTE gradients were designed to sample k-space at 1/r using in-house Python software. The trajectories were designed to have 16380 spokes, TR/TE of 30/0.5 ms, field of view of 240 mm, acquisition time of 8:11. 3D polar coordinate angles were calculated from a set of 16380 points equally distributed on a sphere. Transmit gain and frequency calibrations were performed by using the Bloch-Siegert shift.6
Reconstruction was performed off-line using the non-uniform fast fourier transform (NUFFT) from the Berkeley Advanced Reconstruction Toolbox,7 and then converting reconstructions into NIFTI files, dimensions of 160 x 160 x 160.
Mango (http://ric.uthscsa.edu/mango/) was used for the analysis of the cross-section data across the 9 different phantom resolutions, from which modulation transfer functions were calculated. Data were then linear interpolated onto a 1D grid before calculating normalized PSFs. The PSFs were assumed Gaussian for fitting purposes. Python and Matplotlib8 were used to visualize the data and results. Signal to noise ratios (SNR) were also calculated with Mango -- regions of interest were placed inside the phantom in a homogeneous-appearing region and outside the phantom in incoherent noise.
Results
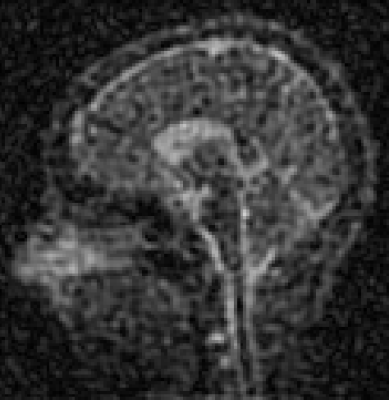
Figure 2 has representative slices from the 3 phantom concentrations. Images are progressively noisier as the concentration of saline decreases in the phantom. Figure 3 displays the calculated PSF data and fitted Gaussian functions for the 3 phantom concentrations. SNR and PSF full-width half maximum results can be found in Table 1. Figure 4 displays a representative sagittal slice from the volunteer.
Discussion
In vivo sodium MRI is challenging because of the scarcity of available signal. As the signal is proportional to the concentration of 23Na, and the performance of the imaging system is correlated with signal (see Figure 3 and Table 1), it is expected that any further reduction of signal will negatively impact image resolution. This work examined performance in a saline phantom, which is akin to unbound sodium in vivo. Bound intracellular sodium is of more use diagnostically, and although techniques exist which are sensitive to intracellular sodium concentration,9,10 these also come with a reduction in signal compared to total sodium concentration. Thus, these techniques will further exacerbate the low imaging signal and impact image resolution. This is evident in the in vivo image (Figure 4) -- lower 23Na concentration in the white and grey matter contribute to reduced image quality as compared to higher signal in the cerebrospinal fluid.
Compressed sensing techniques11 provide a potential opportunity to increase 23Na MRI signal without any associated increase in scan acquisition time, especially with regards to parallel imaging reconstructions. Also of interest are the use of other non-conventional UTE acquisition schemes, such as FLORET12 or 3D cones13 -- the performance of these techniques can be measured with the methods presented here.
Conclusions
This work examined the performance of a 3DPR UTE sequence in the context of varying levels of 23Na concentration in a resolution phantom. The results demonstrate some of the difficulties with sodium imaging, in particular that the reduced signal correlates with a reduction in apparent image resolution. Future research will examine the effectiveness of different pulse sequence trajectories and reconstruction techniques in the resolution phantom.Acknowledgements
Thank you to Josh Bierbrier for the initial design of the phantom.References
1. Rose AM, Valdes R. Understanding the sodium pump and its relevance to disease. Clin Chem 1994;40:1674–1685.
2. Skou JC, Esmann M. The Na, K-ATPase. J Bioenerg Biomembr 1992;24:249–261. doi: 10.1007/BF00768846.
3. Madelin G, Lee JS, Regatte RR, Jerschow A. Sodium MRI: Methods and applications. Prog Nucl Magn Reson Spectrosc 2014;79:14–47. doi: 10.1016/j.pnmrs.2014.02.001.
4.Madelin G, Regatte RR. Biomedical Applications of Sodium MRI In Vivo. J Mag Reson Imag 2013;38:511-529. doi: 10.1002/jmri.24168.
5. Nagel AM, Laun FB, Weber M-A, Matthies C, Semmler W, Schad LR. Sodium MRI using a density-adapted 3D radial acquisition technique. Magn Reson Med 2009;62:1565–1573. doi: 10.1002/mrm.22157.
6. Schulte RF, Sacolick L, Deppe MH, Janich MA, Schwaiger M, Wild JM, et al. Transmit gain calibration for nonproton MR using the Bloch-Siegert shift. NMR in Biomedicine. 2011 Nov 1;24(9):1068-1072.
7. Uecker M, Ong F, Tamir JI, Bahri D, Virtue P, Cheng JY, Zhang T, Lustig M. Annual Meeting ISMRM, Toronto 2015, In Proc Intl Soc Mag Reson Med 23:2486.
8. Hunter J. Matplotlib: A 2D graphics environment. Computing in Science & Engineering. 2007;9(3):90-95.
9. Ooms KJ, Cannella M, Vega AJ, Marcolongo M, Polenova T. 23Na TQF NMR imaging for the study of spinal disc tissue. J Magn Reson. 2008 Nov;195(1):112-115.
10. Stobbe R, Beaulieu C. In vivo sodium magnetic resonance imaging of the human brain using soft inversion recovery fluid attenuation. Magn Reson Med. 2005;54(5):1305-1310.
11. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007 Dec;58(6):1182–95.
12. Robison RK, Anderson AG, Pipe JG. Three-dimensional ultrashort echo-time imaging using a FLORET trajectory. Magn Reson Med. 2017;78(3):1038-1049.
13. Gurney PT, Hargreaves BA, Nishimura DG. Design and analysis of a practical 3D cones trajectory. Magn Reson Med. 2006 Mar;55(3):575-582.
Figures


Figure 2: 23Na MRI reconstructed images for saline phantom at concentrations of 150, 60, and 30 mM (left to right). Images have been windowed identically. The phantom scans were all performed in the coronal plane, and subsequently reformatted to match the CT reconstruction (Figure 1) for comparison purposes. As all 3D acquisitions were isotropic this was permitted.