2400
Feasibility Study of Improving SPIRiT by Exploiting Artificial Sparsity in Dynamic MRI12nd Affiliated Hospital, Zhejiang University, Hangzhou, China, 2Radiology, 2nd Affiliated Hospital, Zhejiang University, Hangzhou, China, 3School of Computer and Information Science, Hubei Engineering University, Xiaogan, China, 4School of Biomedical Engineering, Southern Medical University, Guangzhou, China, 5Guangdong Provincial Key Laboratory of Medical Image Processing, Guangzhou, China
Synopsis
Improving spatiotemporal resolution is of great importance for dynamic MRI in clinical circumstances. An improved SPIRiT method using artificial sparsity and PCA denoising is proposed in this work. Simulated cardiac perfusion phantom and in-vivo cardiac cine experiments were conducted. The proposed method showed better image quality compared with GRAPPA and the frame-by-frame SPIRiT method.
Introduction
High spatial and temporal resolutions are both important for dynamic MRI. However, there is a trade-off between spatial resolution and temporal resolution in dynamic MRI, because of the limitation of the acquisition speed of the MRI scanners. Partially parallel imaging (PPI) methods have been widely used in clinical applications such as SENSE and GRAPPA, which acquire partial k-space data instead of full sampling. SPIRiT is a PPI method which is heavily based on GRAPPA but also similar to SENSE in the sense that the reconstruction is the solution for a least-squares optimization.1 When the reduction factor goes higher, traditional PPI methods encounter problems associated with noise amplification and residual aliasing artifacts.2 It is found that PPI performs better when the image support is artificially reduced, which is called artificial sparsity.2-4 In this work, we propose to extend SPIRiT by including artificial sparsity and principal component analysis (PCA) denoising, termed as ARTS-SPIRiT. The performance of this artificial sparsity method compared to GRAPPA and frame-by-frame SPIRiT reconstruction is evaluated using myocardial perfusion phantom and cardiac cine dataset.Methods
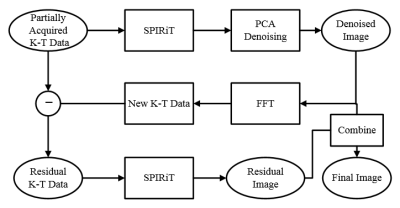
Fig. 1 depicts the flowchart of the proposed ARTS-SPIRiT method: (I) the frame-by-frame SPIRiT method is applied to the undersampled K-T data (K-T data 1); (II) the initial reconstructed image is denoised with PCA method, the denoised image is transformed to K-T data (K-T data 2) via Fast Fourier Transform (FFT); (III) residual K-T data is obtained by complex subtracting K-T data 2 from K-T data 1; (IV) frame-by-frame SPIRiT is then applied to the residual K-T data to generate residual image; (V) final reconstructed image is restored by summing the residual image and the denoised image.Experiments
A simulation experiment was conducted using the numerical cardiac perfusion phantom MRXCAT to evaluate the performance of the proposed ARTS-SPIRiT method.5 The relevant simulation parameters were as follows: repetition time/echo time = 2/1 ms; flip angle= 15∘; spatial resolution = 2 × 2 × 5 mm3, matrix size = 224 ×192, there would be 32 temporal frames collected by four coils, and the normalized distributed white Gaussian noise was added to the phantom. The proposed technique was also verified with an online-accessed two-dimensoional cine (in short axis view of the heart) dataset, which was acquired on a 3.0T scanner (Ingenia, Philips Healthcare, The Netherlands) with a 28-channel coil array. The scan parameters of the balanced SSFP sequence included a field-ofview of 270×270 mm2, 8 mm slice thickness, TR/TE of 3.8/1.84 ms, voxel size of 1.4×1.4 mm2, 45° flip angle, 192 × 190 acquisition matrix, and 23–28 heart phases, the raw data is avaiable at https://osf.io/n89tq/.6 The simulation and in-vivo data were undersampled with reduction factors of 4 and 8, and were reconstructed using GRAPPA, frame-by-frame SPIRiT, and ARTS-SPIRiT respectively.Evaluation Criteria
The absolute error map and normalized root-mean-square error (NRMSE) are used to evaluate the reconstruction results. NRMSE is defined as: $$ \frac{\parallel I \scriptsize ref \normalsize (r)-I(r) \parallel \tiny F}{ \parallel I \scriptsize ref \normalsize (r) \parallel \tiny F} $$, where $$$ I \scriptsize ref \normalsize (r)$$$denotes the reference image; $$$I(r)$$$ is the undersampled reconstruction result, and $$$r$$$ is the spatial location of the image, $$$\parallel I \scriptsize ref \normalsize (r) \parallel \tiny F $$$ is the Frobenius norm of the corresponding image.Results
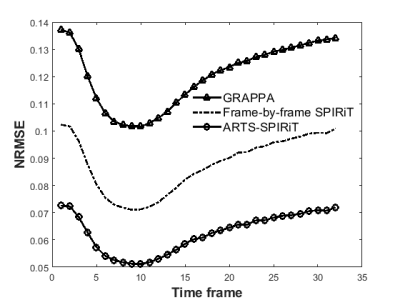
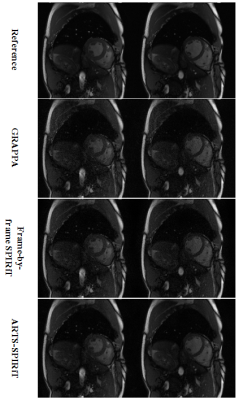
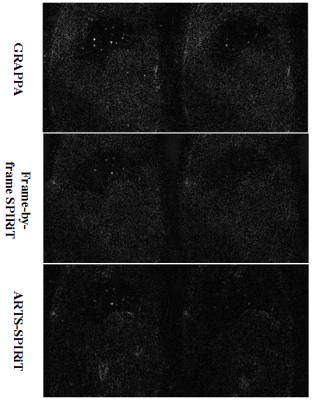
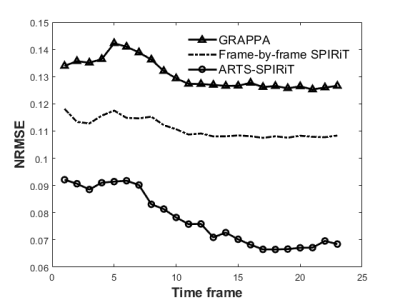
The NRMSE results of the simulation experiment are shown in Fig. 2. It can be seen that the NRMSE of SPIRiT and ARTS-SPIRiT are lower than that of GRAPPA, and ARTS-SPIRiT generates the lowest NRMSE. As for the in-vivo experiment, the reference images and images reconstructed using GRAPPA, frame-by-frame SPIRiT, and ARTS-SPIRiT are all displayed in Fig. 3. Absolute error maps between the reference images and the corresponding reconstructed images are shown in Fig. 4. NRMSE of ARTS-SPIRiT is the lowest, which is shown in Fig. 5.Discussion
This work has presented an improved SPIRiT method using artificial sparsity in dynamic MRI. The image artifacts and the noise caused by high reduction factor were effectively suppressed by adding artificial sparsity and PCA denoising. NRMSEs of the proposed ARTS-SPIRiT method are always the lowest compared with that of GRAPPA and frame-by-frame SPIRiT. In further study, this method will also be validated with in-vivo liver dynamic contrast-enhanced experiments and be compared with other K-T reconstruction schemes such as K-T PCA, K-T SPARSE-SENSE and K-T ARTS-GROWL.2,4,7Conclusion
The ARTS-SPIRiT method to reconstruct dynamic MRI was described and validated with numerical cardiac perfusion phantom and in-vivo cardiac cine dataset. Compared with GRAPPA and frame-by-frame SPIRiT, the proposed method shows better image quality.Acknowledgements
This project is supported in part by the Science Technology Department Program of Zhejiang Province (LGG18H180001) and the Project of Medical and Health Technology Development Program of Zhejiang Province (2019KY081).References
[1] Lustig M and Pauly J M. SPIRiT: Iterative self-consistent parallel imaging reconstruction from arbitrary k-space. Magnetic Resonance in Medicine. 2010;64(2): 457-471.
[2] Wang Y, Chen Z, Wang J, et al. Improved - PCA Algorithm Using Artificial Sparsity in Dynamic MRI. Computational and Mathematical Methods in Medicine. 2017. Article ID 4816024, doi:10.1155/2017/4816024.
[3] Chen Z, Xia L, Liu F, et al. An improved non-Cartesian partially parallel imaging by exploiting artificial sparsity. Magnetic Resonance in Medicine. 2017;78(1): 271–279.
[4] Chen Z, Kang L, Xia L, et al. Sequential combination of parallel imaging and dynamic artificial sparsity framework for rapid free-breathing golden-angle radial dynamic MRI: K-T ARTS-GROWL. Medical Physics. 2018;45(1): 202–213.
[5] Wissmann L, Santelli C, Segars W P, et al. MRXCAT: Realistic numerical phantoms for cardiovascular magnetic resonance. Journal of Cardiovascular Magnetic Resonance. 2014;16:63. https://doi.org/10.1186/s12968-014-0063-3
[6] Schmidt J F, Santelli C, Kozerke S. MR image reconstruction using block matching and adaptive kernel methods. PLOS ONE. 2016; DOI:10.1371/journal.pone.0153736.
[7] Feng L, Srichai M B, Lim R P, et al. Highly accelerated real-time cardiac cine MRI using k-t SPARSE-SENSE. Magnetic Resonance in Medicine. 2013;70(1): 64–74.
Figures