1408
Prospective Fast T1ρ Mapping of Knee Articular Cartilage using Compressed Sensing1Program of Advanced Musculoskeletal Imaging (PAMI), Cleveland Clinic, Cleveland, OH, United States, 2Electrical Engineering, Case Western Reserve University, Cleveland, OH, United States, 3Electrical Enigeering, University of Buffalo, Buffalo, OH, United States
Synopsis
Quantitative magnetic resonance (MR) mapping is known to provide additional information compared to the conventional qualitative weighted MR images. Relaxation times such as spin-spin relaxation time (T2) and spin-lattice relaxation time (T1ρ) have been shown enable early detection of human knee cartilage degeneration. Obtaining these relaxation parameter maps, however, requires long acquisition time. Compressive sensing (CS) has been extensively studied over the last decade as one of the possible ways to accelerate MR acquisition. These studies, however, are retrospective. In this study, we perform a prospective downsampling study on T1ρ mapping of knee cartilage using CS, which implements the downsampling pattern into the in vivo MR scan sequence.
INTRODUCTION
Quantitative magnetic resonance (MR) mapping can provide additional information compared to qualitative MR images. Relaxation times such as spin-spin relaxation time (T2) and spin-lattice relaxation time (T1ρ) have been shown to enable detection early human knee cartilage degeneration before morphological changes appear1. Obtaining these relaxation parameter maps, however, requires long acquisition time. In particular, due to the thin structure of articular cartilage, high resolution maps are desirable. This can introduce patient discomfort, increased motion artifacts, and increased cost.
Compressive sensing (CS) has been extensively studied over the last decade as one way to accelerate MR acquisition2. The feasibility of applying CS in cartilage T1ρ has been shown recently3,4. These studies, however, are retrospective. The acceleration is typically achieved based on the already collected fully sampled k-space data, which brings biases caused by signal to noise ration (SNR), and noise/artifacts pattern. In this study, we perform a prospective downsampling study on T1ρ mapping of knee cartilage using CS, which implements the downsampling pattern into the in vivo MR scan sequence.
METHODS
In vivo human knee data are collected on a 3T MR scanner (General Electric Healthcare, Milwaukee, WI, USA) using a 1Tx/8Rx knee coil (InVivo). Three volunteers were scanned using a magnetization-prepared angle-modulated partitioned k-space spoiled gradient echo snapshots (MAPSS)5 T1ρ quantification pulse sequence with spin-lock times (TSLs) of 0, 2, 4, 8, 12, 20, 40, 80ms, spin-lock frequency 500Hz, matrix size 128×192×8×28 (PE×FE×Echo×Slice), FOV 14cm2, and slice thickness 4mm. Prospectively subsampled k-space data with a variable density random sampling pattern along the phase encoding direction for each echo with acceleration factor 2, 3, 4 were collected.
The collected data were reconstructed by iterating between a CS based dynamic imaging technique, namely, k-tLAISD, and a parallel imaging technique named JSENSE. Specifically, principal component analysis (PCA) was applied to low resolution images obtained from fully sampled k-space center to obtain the sparsifying transform for CS reconstruction. An initial guess of the principal components (PC) was calculated by solve an l1-regularized Lasso problem with combined coil sensitivity maps. Then, local support detection was applied to the PC space to update the PCs and the local supports. The PCs were further updated by solving another l1-minimization problem with restricted sparsity supports. The coil sensitivity maps were further updated by applying JSENSE.
The reconstructed T1ρ-weighted images for different echoes were registered to images with the shortest TSL and highest SNR. Levenberg-Marquardt mono exponential fitting is applied to construct the T1ρ maps. Cartilage were segmented semi-automatically in high-resolution morphologic images (3D fast spin-echo images, FOV 140mm2, Matrix 384×384, slice thickness 1mm) into six compartments including lateral femoral condyle (LFC), lateral tibia (LT), medial femoral condyle (MFC), medial tibia (MT), patella (PAT), trochlea (TRO). The segmentation was overlaid to the T1ρ maps after registration. The mean and standard deviation of T1ρ values for each compartment were then calculated. Coefficients of variation (CVs) were calculated between T1ρ values obtained from fully sampled data and downsampled data.
RESULTS and DISCUSSION
The proposed prospective CS downsampling significantly reduced the acquisition time for T1ρ calculation (Table. 1). Moreover, by applying local support detection, it enables the algorithm to adapt to the SNR of cartilage region, which has relatively high SNR compared to surrounding areas, especially in later TSLs. This in return benefits the T1ρ quantification.
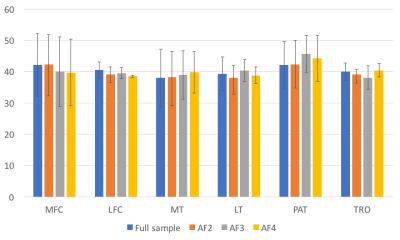
Figure 1 shows the comparison of the T1ρ mean values among the fully sampled data and compressive sensing reconstructed ones with different acceleration factors. As shown in the figure, the T1ρ values fitted from CS reconstructed images agree well with the ones fitted from the fully sampled images for all the six knee cartilage compartments.
Table. 2 further confirms the above findings. It shows the average coefficients of variations (CV) between the T1ρ values fitted using CS reconstructed images and the fully sampled data for all the 6 knee cartilage compartments. In all three volunteers, no significant difference in T1ρ values is found between fully sampled data and subsampled data. All CVs are below 6%, which is comparable with the reported in vivo reproducibility of cartilage T1ρ in literature6,7.
CONCLUSION
This work implemented the prospective subsampling acquisition and reconstruction based on CS and JSENSE to accelerate T1ρ quantification of knee articular cartilage. The CS reconstruction algorithm has the advantage of applying locally adaptive thresholding support detection in terms of SNR and the effectiveness of JSENSE in sensitivity estimation. It reduced the T1ρ quantification acquisition time from 20 minutes to 5 minutes with minimal variation comparable to in vivo reproducibility, which makes clinical T1ρ mapping feasible. With the design of prospective subsampling pattern in multi-dimensions, it is possible to further reduce the acquisition time for reduced cost and improved patient comfort.Acknowledgements
No acknowledgement found.References
1. Li X, Majumdar S. Quantitative MRI of articular cartilage and its clinical applications. J Magn Reson Imaging. 2013;38:991-1008.
2. Lustig M, Donoho DL, Santos JM, Pauly JM. Compressed Sensing MRI. IEEE Signal Processing Magazine. 2008;25(2):72-82.
3. Zhou Y, Pandit P, Pedoia V, et al. Accelerating t1ρ cartilage imaging using compressed sensing with iterative locally adapted support detection and JSENSE. Magn Reson Med. 2016;75:1617-1629.
4. Zibetti MVW, Sharafi A, Otazo R, Regatte RR. Compressed sensing acceleration of biexponential 3D‐T1ρrelaxation mapping of knee cartilage. Magn Reson Med. 2018;00:1–18.
5. Li X, Han E, Busse R, Majumdar S. In vivo T1rho mapping in cartilage using 3D magnetization-prepared angle-modulated partitioned k-space spoiled gradient echo snapshots (3D MAPSS). Magn Reson Med. 2008;59(2):298-307.
6. Li X, Pedoia V, Kumar D, et al. Cartilage T1rho and T2 relaxation times: longitudinal reproducibility and variations using different coils, MR systems and sites. Osteoarthritis Cartilage. 2015;23(12):2214-23.
7. MacKay JW, Low SBL, Smith TO, et al. Systematic review and meta-analysis of the reliability and discriminative validity of cartilage compositional MRI in knee osteoarthritis. Osteoarthritis Cartilage. 2018. Epub 2018/03/20.