1109
Sub-millimeter MR Fingerprinting using deep-learning-based spatially-constrained tissue quantification1Radiology, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 2Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 3Biomedical Engineering, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Synopsis
MRF is a relatively new quantitative MR imaging technique which can provide rapid and simultaneous quantification of multiple tissue properties. However, high-resolution MRF, particularly at sub-millimeter levels, is technically challenging and often requires extended scan time. In this study, a rapid high-resolution MRF technique was developed using a deep-learning-based spatially-constrained tissue quantification method. The experimental results from in vivo brain data demonstrate that high-quality T1 and T2 quantification with 0.8-mm resolution can be achieved in 15 sec per slice.
Target Audience
This work targets those interested in high-resolution quantitative imaging and deep learning.Purpose
MR Fingerprinting (MRF) is a new framework for image acquisition, reconstruction and post-processing, which can provide rapid and simultaneous quantification of multiple tissue properties (1). While MRF has demonstrated better performance as compared to other quantitative techniques for T1 and T2 mapping, further improvement in both spatial resolution and acquisition speed will facilitate wide applications especially for pediatric population and imaging of small organs. However, high-resolution MRF, particularly at sub-millimeter levels, is technically challenging and often requires extended scan time. Recently, a deep-learning-based spatially-constrained tissue quantification (SCQ) method has been developed to estimate tissue properties for MRF (2). Compared to the conventional dictionary matching (DM) approach, this method could extract more useful features from the MRF signal evolutions and provide accurate quantification with highly reduced sampling data. The purpose of this study was to leverage the SCQ method to develop a rapid MRF technique with a sub-millimeter spatial resolution.Methods
All the MRF datasets used in this study were acquired on a Siemens 3T Prisma scanner with a 32-channel head coil. The 2D MRF method previously developed for cardiac MRF was adopted (3) and a spiral trajectory with 0.8 mm resolution was designed and applied in the measurements (FOV, 25 cm; matrix size, 304×304; slice thickness, 3 mm). A total of 2304 time frames were acquired with variable flip angles ranging from 5° to 12°. Instead of acquiring only one spiral arm for each time frame as the conventional MRF approach (reduction factor of 48), four spiral arms were acquired for each time frame (reduction factor of 12) to ensure high-quality quantitative maps for the training purpose. A constant waiting time of 20 sec was applied between sampling of different spiral arms to ensure completely longitudinal relaxation for all brain tissues including cerebrospinal fluid. The total scan time for one slice was ~3 min. Five normal volunteers (M:F, 3:2; mean age, 29±13 years) were recruited for this study and a total of ten slices were acquired for each subject.
The structure of SCQ model used for tissue quantification in this study is shown in Fig. 1. The model contained two modules. First, a feature extraction (FE) module was used to extract features from each signal evolution for dimension reduction. Then, a spatially-constrained quantification (SQ) module was used to estimate tissue property maps from the low-dimensional feature maps. The model was trained by minimizing relative difference between network’s output and ground truth. In each experiment, 40 slices from 4 subjects were selected as training data, and the slices from the remaining one subject were used for testing. Patches of 64x64 pixels were extracted for training. The high-quality T1 and T2 maps obtained from four spiral arms and all 2304 time points by DM were used as ground truth. For fast imaging purpose, the data of only one spiral arm with different numbers of time frames (576, 1152, and 2304) was used as input for the SCQ model. The results were compared with those obtained with the standard DM method.
Results
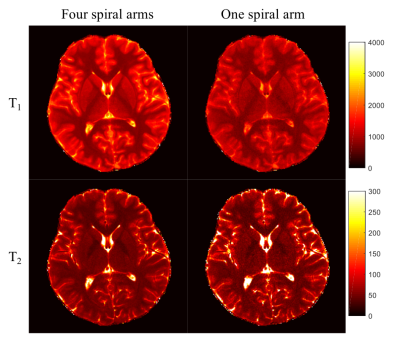
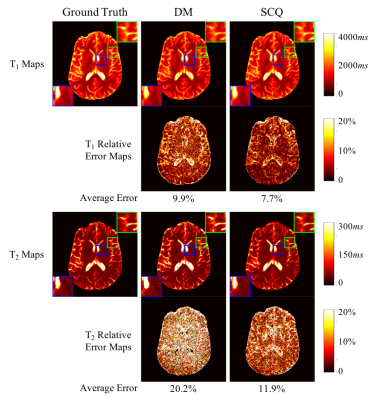
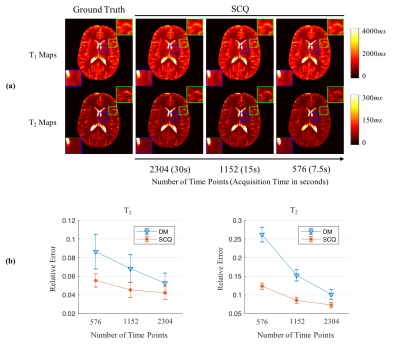
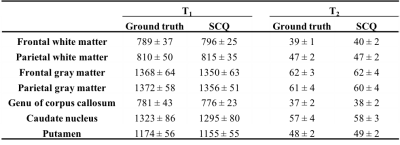
Fig. 2 shows T1 and T2 maps obtained using DM with only one spiral arm and all four spiral arms. With the high spatial resolution of 0.8 mm, the quality of tissue property maps was largely improved with acquisition of more spiral arms, which was selected as the ground truth for training in this study. Fig. 3 shows results from DM and SCQ using one spiral arm and 1152 (50%) time points. The effect of different numbers of time points is shown in Fig. 4. Compared to the standard DM method, SCQ achieved higher quantification accuracy in both T1 and T2 for high-resolution data with one spiral arm. No visual difference was observed in the results between 2304 and 1152 time points, whereas some image blurring was noted in T2 maps with 576 points. Leave-one-out cross validation was further performed using 1152 points, and T1 and T2 values from different brain tissues are summarized in Table 1.Discussion and Conclusion
In this study, a rapid MRF technique with sub-millimeter spatial resolution was developed, and our results show that high-quality T1 and T2 quantification with 0.8-mm resolution can be achieved in 15-sec acquisition time for each slice. This enables reliable tissue quantification of fine structures such as cortical gray matter in brain imaging. Future studies will also be performed to extend the method for other organs in the body such as prostate and cartilage imaging.Acknowledgements
No acknowledgement found.References
1. Ma D, et al. Nature, 2013; 187–192.
2. Fang Z, et al. MLMI 2018, Proceedings; 398-405. DOI: 10.1007/978-3-030-00919-9_46.
3. Hamilton JI, et al. MRM, 2018; 40-51.
4. Ronneberger O, et al. MICCAI, 2015; 234-241.
Figures