0855
Noninvasive Assessment of Tumor Histopathology in Glioblastoma Specimen and Patient1Radiology, Washington University School of Medicine, Saint Louis, MO, United States, 2Neurological Surgery, Washington University School of Medicine, Saint Louis, MO, United States, 3Medical Scientist Training Program, The University of Alabama at Birmingham, Birmingham, AL, United States, 4Medicine, University of Missouri – Kansas City, Kansas City, MO, United States, 5Radiology, The First Affiliated Hospital of Nanchang University, Nanchang, China, 6Radiology, Guangzhou First People’s Hospital, Guangzhou, China, 7Pathology and Immunology, Washington University School of Medicine, Saint Louis, MO, United States
Synopsis
Glioblastoma (GBM) is the most frequent malignant brain tumor in adults, accounting for approximately 45-50% of all primary malignant brain tumors. To circumvent the shortcomings of current clinical MRI techniques, we applied modified diffusion basis spectrum imaging (DBSI) to accurately detect pathology in GBM. In this study, we demonstrate modified-DBSI efficacy in detecting different histopathological structures in ex vivo and in vivo scans of glioblastoma.
Introduction
Glioblastoma (GBM) is the most frequent malignant brain tumor in adults, accounting for approximately 45-50% of all primary malignant brain tumors. The presence of highly anaplastic glial cells, mitotic activity, and microvascular proliferation and/or necrosis is required for pathological diagnosis, but regional heterogeneity of GBM is significant. For GBM, gadolinium (Gd)-enhanced T1-weighted MRI is the current standard for diagnosis, surgical planning, and evaluating treatment response. Gd-T1W imaging, however, is not specific for glioblastoma, is unable to detect tumor infiltration beyond the edges of enhancing regions, and cannot distinguish true tumor progression vs. pseudoprogression. Diffusion MRI-derived ADC is another imaging technique used to assess tumor cellularity, but is limited due to the complicated GBM microenvironment. To circumvent the shortcomings of current clinical MRI techniques, we aimed to use modified diffusion basis spectrum imaging (DBSI) to accurately detect pathology in GBM. In this study, we demonstrate modified-DBSI efficacy in detecting different histopathological structures in ex vivo and in vivo scans of glioblastoma.Materials and Methods
Twenty brain tumor specimens from fourteen patients with glioblastoma were surgically resected and immediately fixed in 10% paraformaldehyde. Specimens were examined using a 4.7T Agilent MR scanner and a home-made surface coil. A spin-echo diffusion-weighted sequence with 99 diffusion encoding directions and maximum b-value=3000 s/mm2 was employed to acquire DW images with the following imaging parameters: TR=1500 ms, TE=40 ms, slice thickness=0.5 mm, resolution=0.25×0.25 mm2. A 3 T Siemens TIM Trio scanner with a 32-channel head coil was used for all in vivo studies. Diffusion weighted MRI data with 99-direction diffusion-encoding-scheme and maximum b-value=1500 s/mm2 were collected with a 2x2x2 mm3 resolution in the axial plane covering the whole brain. Tissues were embedded in agar gel for MR imaging followed by sectioning for H&E. H&E images were reviewed by an experienced neuropathologist. DBSI maps were linearly co-registered with corresponding H&E images, allowing MRI voxels to be labeled by the histological gold standard.Results
In a specimen from a 77-year old female patient with GBM, the region of increased cellularity detected by DWI hyperintensity and ADC hypointensity (Fig. 1A) did not match the H&E staining (Fig. 1B). On the other hand, regions of dense tumor, necrosis and white matter infiltration exhibited specific modified-DBSI metric profiles corresponding to individual pathological regions (Fig 1C).
After co-registration between MRI and H&E staining, 7800 image voxels of diffusion data from 20 GBM specimens were gathered to compare profiles of DTI and modified-DBSI metrics (Fig. 2). DTI-ADC was highest in dense tumor, contradicting utility as a marker of increased tumor cellularity. DTI-FA was highly overlapping among all three pathologies. Modified-DBSI restricted fraction was highest for dense tumors; hindered fraction was highest for necrosis; and anisotropic fraction was lowest in dense tumor among all three pathologies. Modified-DBSI derived diffusion anisotropy, restricted and hindered isotropic diffusion fractions correctly separated pathologies (Fig. 2B).
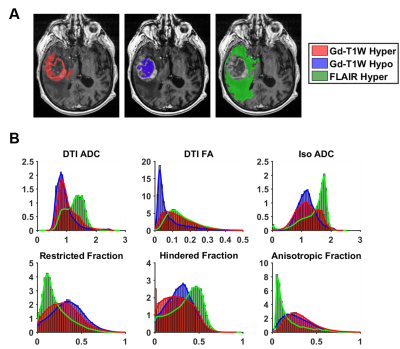
Modified-DBSI analysis was performed and compared to multiparametric MRI of a newly diagnosed GBM patient (Fig. 3). Areas of high cellularity suggested by modified-DBSI restricted fraction did not correspond closely to Gd-enhancing areas (Fig. 3, white arrows). In contrast, modified-DBSI predicted a large region of hyper-cellularity within the non-Gd-enhancing tumor core (Fig. 3, yellow arrows). Three dimensional modified-DBSI reconstruction illustrated distribution of tumor cellularity and necrosis in contrast to that of Gd-enhancement (Fig. 4).
We generated modified-DBSI restricted-, and hindered-diffusion-fraction distributions for Gd-enhancing, non-enhancing, and FLAIR hyperintense voxels. Remarkably, Gd-enhancing and non-enhancing voxels appear equally likely to be represented in areas of high restricted-diffusion, or high cellularity (Fig. 5B). Similarly, the distribution of hindered-fraction suggests that necrosis can be found in both Gd-enhancing and non-enhancing voxels. We detected similar distributions between Gd-enhancing and Gd-non-enhancing regions on DTI-ADC, consistent with findings from the modified-DBSI map. In FLAIR hyperintense voxels, the restricted-fraction suggested a generally lower burden of cellularity. This, along with the low modified-DBSI anisotropic-fraction and low DTI-FA, suggests disruption of white matter, potentially resulting from infiltrating tumor cells or edema.
Discussions and Conclusions
Accurate assessment of tumor cellularity, necrosis, and tumor infiltration is critical for clinical management of GBM patients. However, the spatial distribution of these tumor pathologies cannot be easily assessed via traditional MRI methods. Through voxel-wise comparisons of histological images, we showed that modified-DBSI derived restricted-diffusion-fraction, hindered-diffusion-fraction, and anisotropic-diffusion-fraction correlated with tumor cellularity, necrosis, and white matter tracts, respectively. We demonstrated that modified-DBSI could guide preoperative evaluation to better facilitate clinical decision-making without imposing unnecessary risk and discomfort to patients. Ultimately, this method may help to better select regions for biopsy and diagnosis and potentially improve maximal safe extent of resection based on tumor cellularity.Acknowledgements
This work was supported in part by NIH R01-NS047592, P01-NS059560, U01-EY025500, National Multiple Sclerosis Society (NMSS) RG 5258-A-5, RG 1701-26617, and Department of Defense Idea Award W81XWH-12-1-0457, the Fundamental Research Funds for the Central Universities, SCUT (2018MS23), and Natural Science Foundation of Guangdong Province in China (2018A030313282).References
1. Yamasaki F, Kurisu K, Satoh K, et al. Apparent diffusion coefficient of human brain tumors at MR imaging. Radiology. Jun 2005;235(3):985-991.
2. Wang Y, Wang Q, Haldar JP, et al. Quantification of increased cellularity during inflammatory demyelination. Brain : a journal of neurology. Dec 2011;134(Pt 12):3590-3601.
Figures